Full Articles
A mini-review of current understanding of the Hedgehog signaling pathway
Contents:
- Introduction
- Hedgehog signaling cascade
- Processing of Hedgehog ligand
- Hedgehog signaling orthologues in vertebrates
- References
Introduction
The Hedgehog gene was identified in genetic screens aimed to provide an understanding of body segmentation in Drosophila (Nusslein-Volhard et al., 1980). Loss of the secreted Hedgehog signaling protein was found to cause Drosophila embryos to develop as spiny balls reminiscent of hedgehogs. Hedgehog signaling plays a role in many processes during embryonic development and remains active in the adult where it is involved in the maintenance of stem cell populations. Here, aberrant Hedgehog signaling in some cases can lead to certain forms of cancer.
Hedgehog signaling cascade
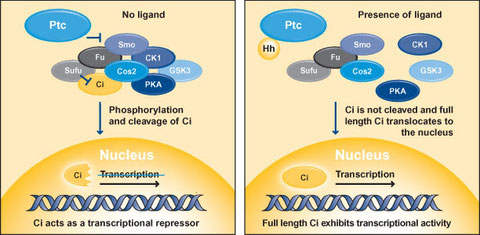
In Drosophila, Hedgehog signaling is initiated by the binding of Hedgehog ligand to Patched (Ptc) (Figure 1), which is a 12-transmembrane protein receptor (Cohen 2003; Ogden et al., 2004; Hooper and Scott 2005; Østerlund and Kogerman 2006; Jia and Jiang 2006; Huangfu and Anderson 2006). Ptc acts as an inhibitor of Smoothened (Smo), a 7-transmembrane protein related to the Frizzled family of Wnt receptors and to other 7-transmembrane G protein-coupled receptors. Downstream of Smo is a multi-protein complex known as the Hedgehog signaling complex (HSC), which comprises the transcription factor Cubitus interruptus (Ci), the serine/threonine kinase Fused (Fu), the kinesin-like molecule Costal 2 (Cos2) and Supressor of fused (Sufu). Cos2 also binds to protein kinase A (PKA), protein kinase CK1 (formerly casein kinase 1) and glycogen synthase kinase 3 (GSK3), which are other kinases that are implicated in the Hedgehog signaling pathway.
In the absence of ligand, Ptc represses Smo preventing the activation of Hedgehog signaling (Hooper and Scott 2005; Østerlund and Kogerman 2006). The HSC is bound to microtubules/membranes and associates with Smo through Cos2. The full length form of Ci is prevented from nuclear translocation through interactions with Sufu and Cos2. A portion of full length Ci is proteolytically cleaved to produce a repressor form of Ci, which enters the nucleus leading to the inhibition of Hedgehog target gene expression. Proteolytic processing of Ci is mediated by PKA, CK1 and GSK3. In the presence of Hedgehog, the inhibitory affects of Ptc on Smo are relieved and the HSC is freed from microtubules and membranes. Smo becomes phosphorylated by PKA and CK1 and PKA, CK1 and GSK3 are released from Cos2, precluding the generation of the repressor from of Ci. Full length Ci is no longer inhibited by Sufu and is therefore free to enter the nucleus to induce the transcription of Hedgehog target genes such as engrailed, Ptc and decapentaplegic (encodes Bone Morphogenetic Proteins in vertebrates).
Processing of Hedgehog ligand
Hedgehog is synthesized as a precursor, which undergoes an autoproteolytic cleavage to liberate a 19 kDa N-terminal fragment (N-Hh), which displays all known signaling properties and a slightly larger C-terminal peptide fragment that has no apparent function other than to catalyse cleavage (Murone et al., 1999; Mullor et al., 2002). The auto-processing reaction provides a trigger for the addition of a cholesterol moiety to the C-terminal of N-Hh. The N terminus is modified via the addition of a palmitate molecule by the acyltransferase Skinny Hedghog (Skn) - also known as Central missing (Cmn), Rasp and Sightless. Membrane tethered Hedgehog proteins initiate signaling in the nearby vicinity of the producing cell or Hedgehog proteins form multimeric complexes in which the hydrophobic moieties cluster together in an inner core allowing diffusion of ligand and long range signaling. Dispatched (Disp) and Tout-velu (Ttv) mediate Hedgehog release and diffusion. Disp is required for release of membrane anchored Hedgehog protein and Ttv regulates the synthesis of proteoglycans enabling the movement of Hedgehog ligands thereby facilitating long range signaling.
Hedgehog signaling orthologues in vertebrates
The Hedgehog signaling pathways in vertebrates shares many common features with Drosophila Hedgehog signaling, although distinct differences are also apparent (Hooper and Scott 2005). In mammals there are three Hedgehog genes, Sonic, Indian and Desert Hedgehog. There are also two Ptc genes (Ptc 1 and Ptc 2) as well as three Ci homologues known as Gli 1, Gli 2 and Gli 3. Gli1 and Gli2 are transcriptional activators, whereas Gli 3 functions as a transcriptional repressor. Regulators of Hedgehog signaling in vertebrates include megalin, which is a member of the low-density lipoprotein receptor related family and binds Hedgehog (McCarthy et al., 2002) and SIL which functions downstream of Ptc (Izraeli et al., 2001). Missing in metastasis (MIM or BEG4) is an actin-binding protein that regulates Gli-dependent transcriptional activation in vertebrates, thereby modulating Hedgehog signaling (Callahan et al., 2004).
By Dr Claudie Hooper *
References
Callahan C. A., Ofstad T., Horng L., Wang J. K., Zhen H. H., Coulombe P. A., Oro A. E. (2004) MIM/BEG4, a Sonic Hedgehog-responsive gene that promotes Gli-dependent transcription. Genes Dev. 18, 2724-2729.
Cohen M. M Jr. (2003) The Hedgehog signalling network. Am. J. Med. Genet. 123A, 5-28.
Huangfu D., Anderson K. V. (2006) Signalling from Smo to Ci/Gli: Hedgehog pathways
from Drosophila to vertebrates. Development. 133, 3-14.
Hooper J. E. Scott M . P. (2005) Communicating with Hedgehogs. Nat. Rev. Mol. Cell Biol. 6, 306-317.
Izraeli S., Lowe L.A., Bertness V. L., Campaner S., Hahn H., Kirsch I. R., Kuehn M. R. (2001) Genetic evidence that Sil is required for the Sonic Hedgehog response pathway. Genesis. 31, 72-77.
Jia J., Jiang J. (2006) Decoding the Hedgehog signal in animal development. Cell Mol. Life Sci. 63, 1249-1265.
McCarthy R. A., Barth J. L., Chintalapudi M. R., Knaak C., Argraves W. S. (2002) Megalin functions as an endocytic sonic hedgehog receptor. J Biol. Chem. 277, 25660-25667.
Mullor J. L., Sanchez P., Altaba A. R I. (2002) Pathways and consequences: Hedgehog signalling in human disease. Trends Cell Biol. 12, 562-569.
Murone M., Rosenthal A., de Sauvage F. J. (1999) Hedgehog signal transduction: From flies to vertebrates. Exp. Cell Res. 253, 25-33.
Nusslein-Volhard C., Wieschaus E. (1980) Mutations affecting segment number and polarity in Drosophila. Nature. 287, 795-801.
Østerlund T., Kogerman P. (2006) Hedgehog signalling: how to get from Smo to Ci and Gli. Trends in Cell. Biol. 16, 176-180.
Ogden S. K., Ascano M. Jr., Stegman M. A., Robbins D. J. (2004) Regulation of Hedgehog signalling: a complex story. Biochem. Pharmacol. 67, 805-814.