Molecular Biology News
Visualisation of the Molecular Gateway Across and Into Cellular Membranes

All living organisms are made up of cells, behind these intricate life forms lie complex cellular processes that allow our bodies to function. Researchers working on protein secretion -- a fundamental process in biology -- have revealed how protein channels in the membrane are activated by special signals contained in proteins destined for secretion. The results help explain the underlying mechanism responsible for the release of proteins such as hormones and antibodies into the blood stream.
The findings, published Jan. 25 in the inaugural issue of Cell Reports, represent a major step forward in cell biology. Until now, scientists have been frustrated by not knowing the architecture of the protein transport machinery when engaged by cargo. However, the team led by researchers from the University of Bristol as part of an international collaboration, has successfully produced and visualised such a complex.
All cells are surrounded by membranes, made up from a double layer of fatty molecules called phospholipids. These act as an ideal 'skin', keeping the cell's insides in. In the absence of other components these fatty molecules act as barriers, preventing the necessary rapid exchange of nutrients and waste products, and of larger molecules like proteins, between the environment and the cell interior. However, such movement is required for many proteins to perform their biological functions either within the membrane or the outside.
To overcome this problem, biological membranes contain a number of translocation systems that enable proteins and other useful substances to pass across the phospholipid barrier. In the case of proteins, those destined for transport are recognised by translocation systems via signals embedded in the sequence of amino acids from which they are constructed. Correct passage through or across the membrane is critical in ensuring that cells complete their lifecycle and fulfill their function.
Using electron microscopy and results from X-ray crystallography, Ian Collinson, Professor of Biochemistry at the University, and his team have described the structure of the ubiquitous Sec-complex associated with a bona fide mimic of a pre-secretory protein in the native environment of the membrane. These results reveal how the binding of the signal sequence unlocks the Sec-complex prior to channel opening and pre-protein transport.
Professor Collinson from the University's School of Biochemistry, said: "These findings are important as they address outstanding questions in one of the central pillars of biology, a process essential in every cell in every organism. The results may suggest ways in which the process can be corrupted in order to manage specific disease states or bacteria infections."
University of Bristol. "Scientists map one of life's molecular mysteries: Visualisation of the molecular gateway across and into cellular membranes." ScienceDaily, 26 Jan. 2012. Web. 29 Jan. 2012.
Membrane Fusion A Mystery No More
The many factors that contribute to how cells communicate and function at the most basic level are still not fully understood, but researchers at Baylor College of Medicine have uncovered a mechanism that helps explain how intracellular membranes fuse, and in the process, created a new physiological membrane fusion model.
The findings appear in the current edition of the journal PLoS Biology.
"Within our cells, we have communicating compartments called vesicles (a bubble-like membrane structure that stores and transports cellular products)," said Dr. Christopher Peters, assistant professor of biochemistry and molecular biology at BCM and lead author on the study. "These vesicles migrate through the cell, meet other vesicles and fuse. That fusion process is, in part, mediated through SNARE proteins that bring the vesicles together. How this happens has been in question for years."
The classic model for this process has been studied using artificial liposome models created in a lab. Peters and his colleagues knew a more physiological fusion model had to be studied in order to see a more accurate account of exactly what acts on this process. Using purified yeast organelles they were able to see that more factors come into play than had been originally believed.
In the classic model, it was believed SNARE proteins originating from two opposing membranes are somehow activated and separated into single proteins. Accepter SNARE proteins then form, allowing fusion with another vesicle membrane. How this mechanistically happens has been unknown.
"What we found with our physiological model is that a tethering complex (termed HOPS) is interacting with the SNARE proteins, activating them to begin this process. Also, the SNARE proteins do not completely separate into single proteins as first believed. Only one protein is detached, leaving behind the acceptor complex," Peters said. "This new acceptor SNARE-complex incorporates the single SNARE that has separated from another vesicle and the two vesicles are in position to fuse."
Researchers found that when this tethering factor was removed, the SNARE proteins were unstable and there was no fusion.
"This finding deals with one of the most fundamental reactions in a cell, how membranes fuse with each other. It is important to understand how this works, because when these events go wrong, either accelerating or slowing down, then it can affect certain disorders such as tumor formation," Peters said. "By using our physiological yeast fusion model, the impact of these tethering factors on the SNARE topology can be investigated, along with the many other factors that come into play. This was not the case in the artificial liposome models used in the past."
Source: Baylor College of Medicine
New Clue to Chemical Origins of Life (TNA----- DNA, RNA)
Organic chemists at the University of York have made a significant advance towards establishing the origin of the carbohydrates (sugars) that form the building blocks of life.
A team led by Dr Paul Clarke in the Department of Chemistry at York has re-created a process which could have occurred in the prebiotic world.
Working with colleagues at the University of Nottingham, they have made the first step towards showing how simple sugars -- threose and erythrose -- developed. The research is published in Organic & Biomolecular Chemistry.
All biological molecules have an ability to exist as left-handed forms or right-handed forms. All sugars in biology are made up of the right-handed form of molecules and yet all the amino acids that make up the peptides and proteins are made up of the left-handed form.
The researchers found using simple left-handed amino acids to catalyse the formation of sugars resulted in the production of predominately right-handed form of sugars. It could explain how carbohydrates originated and why the right-handed form dominates in nature.
Dr Clarke said: "There are a lot of fundamental questions about the origins of life and many people think they are questions about biology. But for life to have evolved, you have to have a moment when non-living things become living -- everything up to that point is chemistry.
"We are trying to understand the chemical origins of life. One of the interesting questions is where carbohydrates come from because they are the building blocks of DNA and RNA. What we have achieved is the first step on that pathway to show how simple sugars -- threose and erythrose -- originated. We generated these sugars from a very simple set of materials that most scientists believe were around at the time that life began."
University of York. "Scientists discover new clue to chemical origins of life." ScienceDaily, 24 Jan. 2012. Web. 26 Jan. 2012.
Nanoparticles For More Accurate Delivery Of Cancer Drugs

A new class of nanoparticles, synthesized by a UC Davis research team to prevent premature drug release, holds promise for greater accuracy and effectiveness in delivering cancer drugs to tumors. The work is published in the current issue of Angewandte Chemie, a leading international chemistry journal.
In their paper, featured on the inside back cover of the journal, Kit Lam, professor and chair of the Department of Biochemistry and Molecular Medicine, and his team report on the synthesis of a novel class of micelles called dual-responsive boronate cross-linked micelles (BCMs) , which produce physicochemical changes in response to specific triggers.
A micelle is an aggregate of surfactant molecules dispersed in water-based liquid such as saline. Micelles are nano-sized, measuring about 25-50 nanometers (one nanometer is one billionth of a meter), and can function as nanocarriers for drug delivery.
BCMs are a unique type of micelle, which releases the payload quickly when triggered by the acidic micro-environment of the tumor or when exposed to an intravenously administered chemical compound such as mannitol, an FDA-approved sugar compound often used as a diuretic agent, which interferes with the cross-linked micelles.
"This use of reversibly cross-linked targeting micellar nanocarriers to deliver anti-cancer drugs helps prevent premature drug release during circulation and ensures delivery of high concentrations of drugs to the tumor site," said first author Yuanpei Li, a postdoctoral fellow in Lam's laboratory who created the novel nanoparticle with Lam. "It holds great promise for a significant improvement in cancer therapy."
Stimuli-responsive nanoparticles are gaining considerable attention in the field of drug delivery due to their ability to transform in response to specific triggers. Among these nanoparticles, stimuli-responsive cross-linked micelles (SCMs) represent a versatile nanocarrier system for tumor-targeting drug delivery.
Too often, nanoparticles release drugs prematurely and miss their target. SCMs can better retain the encapsulated drug and minimize its premature release while circulating in the blood pool. The introduction of environmentally sensitive cross-linkers makes these micelles responsive to the local environment of the tumor. In these instances, the payload drug is released primarily in the cancerous tissue.
The dual-responsive boronate cross-linked micelles that Lam's team has developed represent an even smarter second generation of SCMs able to respond to multiple stimuli as tools for accomplishing the multi-stage delivery of drugs to the complex in vivo tumor micro-environment. These BCMs deliver drugs based on the self-assembly of boronic acid-containing polymers and catechol-containing polymers, both of which make these micelles unusually sensitive to changes in the pH of the environment. The team has optimized the stability of the resulting boronate cross-linked micelles as well as their stimuli-response to acidic pH and mannitol.
This novel nano-carrier platform shows great promise for drug delivery that minimizes premature drug release and can release the drug on demand within the acidic tumor micro-environment or in the acidic cellular compartments when taken in by the target tumor cells. It also can be induced to release the drug through the intravenous administration of mannitol.
Source: University of California - Davis
Mutation in 1 copy of U2 snRNA Causing Neurodegeneration
A Jackson Laboratory research team led by Professor and Howard Hughes Medical Investigator Susan Ackerman, Ph.D., has discovered a defect in the RNA splicing process in neurons that may contribute to neurological disease.
The researchers found that a mutation in just one of the many copies of a gene known as U2 snRNAs, which is involved in the intricate processing of protein-encoding RNAs, causes neurodegeneration.
Many so-called non-coding RNAs—those that don't directly encode proteins—are found in multiple copies in the genome, Ackerman says. "These copies are identical, or nearly identical, so conventional wisdom suggested they were redundant. For the first time, we show that a mutation in one copy can lead to disease."
The results, published in the journal Cell, suggest that disease-causing mutations may exist among other repetitive genes. "This opens up a whole new way of studying these RNAs," Ackerman notes, "including the types of disruptions in RNA processing that can lead to degeneration."
Neurons, like most other cells, build the workhorse proteins that carry out vital functions from the genetic "blueprint" encoded in DNA. In broad strokes, DNA gets copied by pre-messenger RNA (pre-mRNA), then pre-mRNA undergoes a splicing process before transporting the genetic code to the ribosome, where proteins are manufactured. But there's much more to it than that.
Specialized RNAs called U-snRNAs are essential to the splicing process. U-snRNAs are highly conserved, meaning that they are found all along the evolutionary pathway from simple organisms to humans. Ackerman showed that mutations in one form of snRNA, known as U2, lead to movement problems and early neuron death in mice.
U2 is a repetitive gene, meaning there are many copies of the same sequence. A mutation in just one copy led to the observed disorders by disrupting alternative splicing events, part of the splicing process that normally allows the creation of two or more protein forms from the same stretch of pre-mRNA.
The error leads to production of mRNAs containing regions known as introns that should have been removed. These abnormal mRNAs cause cell death, either through active toxicity or the production of dysfunctional proteins. Moreover, the researchers noted that the severity of the splicing abnormalities and cell death depend on the "dosage" level of the mutant gene.
Also, Ackerman and her lab noted that highest levels of the mutant U2 were found in the cerebellum, indicating that the expression of mammalian U2s, previously thought to be universal, may be different among various cell types.
Source: Jackson Laboratory
Polar Growth at the Bacterial Scale Reveals Potential New Targets for Antibiotic Therapy
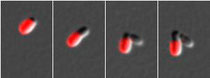
An international team of microbiologists led by Indiana University researchers has identified a new bacterial growth process -- one that occurs at a single end or pole of the cell instead of uniform, dispersed growth along the long axis of the cell -- that could have implications in the development of new antibacterial strategies.
Outer membrane proteins of an Agrobacterium tumefaciens cell were labeled in red, with images taken every 50 minutes as the cell grew. In panels three and four it is clear that the cell on the left (red) has kept all the labeled proteins, whereas the other cell has all new surface proteins.
Based on past detailed studies of rod-shaped bacteria such asEscherichia coli and Bacillus subtilis, it has been assumed that most bacteria grow by binary fission, a dispersed mode of growth involving insertion of new cell wall material uniformly along the long axis of the cell. Growth requires breaking the cell wall at numerous places along the cylinder to allow insertion of new cell wall material, enabling uniform elongation of the cell, with the process culminated by cleavage at the mid-point of the cell to create two symmetric new cells.
The new research published Jan. 17, in Proceedings of the National Academy of Sciences reports on the surprising discovery that cell growth in a large group of rod-shaped bacteria occurs by insertion of new cell wall material only at a single end, or pole, of the cell rather than by the dispersed mode of growth. The cell wall of the progenitor cell remains largely intact, and all of the new cell wall material is partitioned into the new cell.
Polar growth of four bacterial species -- the plant symbiontSinorhizobium meliloti, the plant pathogen Agrobacterium tumefaciens and the human pathogens Brucella abortus andOchrobactrum anthropi -- was observed using time-lapse microscopy and transmission electron microscopy. The four related bacteria used in the study are all members of a large and diverse class of bacteria called the Alphaproteobacteria. The results reported suggest that polar growth is broadly distributed among many different bacterial taxa, including groups outside the Alphaproteobacteria.
There could be a number of reasons why polar growth emerged and has remained conserved and persistent in bacteria, the researchers believe. The process may act as an aid in anchoring damaged material to only the aging mother cell; it could serve as a tool for conservation of energy by constraining growth to a single region of the cell; and ensuring that newborn cells are composed of newly synthesized outer membrane proteins may help pathogens avoid detection by host immune systems.
"As a consequence of polar growth, the two bacterial cells are actually markedly different," said lead author Yves Brun, the Clyde Culbertson Professor of Biology in IU Bloomington's College of Arts and Sciences. "One cell contains all of the old cell wall and surface molecules, including those with damage. In contrast, the other cell is composed of newly synthesized, relatively pristine material."
Ensuring that some cells are composed of newly synthesized surface molecules may help bacteria vary their surface composition, and the ability to do so rapidly is thought to be advantageous for adapting to new environments. Since the defense systems of many plant and animal hosts recognize bacterial cell surfaces, rapid modification of the cell surface may allow bacteria like those used in the experiments to evade detection by the host cell's defense systems.
"These findings make it abundantly clear that the widely accepted binary fission model is not a general rule and suggest that polar growth may be broadly distributed," said IU biology professor Clay Fuqua, one of the IU co-authors. "Therefore, future work aimed at understanding the molecular mechanism underlying polar growth should provide attractive targets for the development of new antibacterial strategies."
Understanding the mechanisms of bacterial growth has enabled advances in strategies to limit the proliferation of bacteria that cause disease. Penicillin, for example, targets actively growing cells by directly inhibiting the proteins responsible for the synthesis of the cell wall and that are required for cell growth. New insights into bacterial cell growth have also been utilized to promote growth of certain bacteria used in oil spill remediation and eradicating disease-carrying mosquitoes.
Co-authors with Brun and Fuqua on "Polar growth in theAlphaproteobacterial order Rhizobiales" included IU Department of Biology postdoctoral researchers Pamela J. B. Brown and David T. Kysela; former IU postdoctoral researcher Jinwoo Kim, who is now at Gyeongsang National University, Korea; Miguel A. de Pedro of the Universidad Autonoma de Madrid, Spain; and Charles Van der Henst and Xavier De Bolle, from University of Namur, Belgium.
Researchers from IU received support for this work from the National Institutes of Health, the National Science Foundation, the IU Metabolomics and Cytomics Initiative and the Korean Research Foundation.
Indiana University. "Polar growth at the bacterial scale reveals potential new targets for antibiotic therapy." ScienceDaily, 17 Jan. 2012. Web. 21 Jan. 2012.
Elusive Z- DNA Found On Nucleosomes

New research published in BioMed Central's open access journal Cell & Bioscience is the first to show that left-handed Z-DNA, normally only found at sites where DNA is being copied, can also form on nucleosomes.
The structure of DNA which provides the blueprint for life has famously been described as a double helix. To save space inside the nucleus, DNA is tightly wound around proteins to form nucleosomes which are then further wound and compacted into chromatin, which is further compacted into chromosomes.
But this familiar image of a right handed coil (also called B-DNA) is not the only form of DNA. At sites where DNA is being copied into RNA (the messenger which is used as the instruction to make proteins) the DNA needs to unwind, and, in a process of negative supercoiling, can form a left-handed variety of the DNA double helix (Z-DNA).
It was originally thought that Z-DNA could only be formed in the presence of active RNA polymerase (the enzyme which assembles RNA). However more recently it has been discovered that SWI/SNF, a protein involved in remodeling nucleosomes and allowing RNA polymerase access to DNA, can convert certain sequences of B to Z-DNA.
The team of researchers led by Dr Keji Zhao discovered that they could convert B-DNA to Z-DNA on nucleosomes by the addition of SWI/SNF and ATP (the cell's energy source) and that the Z-nucleosome formed was a novel structure.
Dr Zhao, from the NIH, explained, "The fact that we have found Z-DNA on nucleosomes is a new step in understanding the roles of chromosome remodeling and Z-DNA in regulating gene expression. While the Z-nucleosome is likely to be a transient structure it nevertheless provides a window of opportunity for the placement of DNA binding proteins which may recruit, regulate, or block the transcription machinery and hence protein expression."
Source: BioMed Central
Gender Differences In Liver Cancer Risk Explained By Small Changes In Genome

Men are four times more likely to develop liver cancer compared to women, a difference attributed to the sex hormones androgen and estrogen. Although this gender difference has been known for a long time, the molecular mechanisms by which estrogens prevent -- and androgens promote -- liver cancer remain unclear.
Now, new research, published in Cell this week from the lab of Klaus Kaestner, PhD, professor of Genetics in the Perelman School of Medicine at the University of Pennsylvania, has found that the difference depends on which proteins the sex hormones bind next to. Specifically a group of transcriptional regulatory proteins called Foxa 1 and 2.
Normally, when mice are given a liver carcinogen, male mice develop many tumors while females get very few. Strikingly, this gender-related incidence of liver cancer was completely reversed in mice genetically engineered by the team to lack the Foxa genes after the team induced cancer. Using complex genomic analyses, the researchers could show that the actions of both estrogens and androgens in the liver are Foxa dependent, explaining the reversal in cancer risk.
But how does this translate to human liver cancer when there are 5,000 places in the human genome where Foxa factors can bind? The team looked for genetic markers called SNPs that intersect with Foxa protein binding. A SNP is a DNA sequence variation occurring when a single nucleotide, or DNA building block, differs between members of a biological species or paired chromosomes in an individual. Knowing that in women the estrogen receptor protects against liver cancer, they looked for SNP markers within Foxa binding sites in tissue samples from women with and without liver cancer.
Strikingly, women with liver cancer frequently had SNPs within specific Foxa binding sites. The researchers then showed that the mutated SNP acts not only to abolish binding of the Foxa proteins, but also of the estrogen receptor to its target sites nearby. This impairment of estrogen receptor binding is thought to result in loss of the protective effect of estrogens, and increased liver cancer risk. Future research will have to determine if the same holds true in reverse in men. In addition, if the human data are validated in larger cohorts of patients, this research might lead to tests for predicting the genetic risk of liver cancer.
Source: University of Pennsylvania School of Medicine
Why Cholesterol-Lowering Statins Might Treat Cancer

Cholesterol-lowering statins seem to keep breast cancer at bay in some patients. Now researchers reporting in the January 20th issue of the journalCell, a Cell Press publication, provide clues about how statins might yield those unexpected benefits. The findings also suggest that mutations in a single gene could be used to identify tumors likely to respond to statin therapy.
The data raises the possibility that we might identify subsets of patients whose tumors may respond to statins," said Carol Prives of Columbia University. "Of course we can't make any definitive conclusions until we know more."
Prives said that a clinical trial of statins in breast cancer based on the mutation status of the tumor suppressor, p53, may be in order. The p53 tumor suppressor acts to regulate many aspects of cell proliferation, generally putting the brake on uncontrolled growth.
More than half of all human cancers carry mutations in the p53 gene. Many of these mutations don't simply disrupt the normal function of p53, they also endow p53 with new functions that promote, instead of inhibit, cancer formation. Mice lacking p53 develop cancer and die, Prives explained, but mice carrying tumor-derived mutant forms of the p53 gene suffer from more aggressive disease. What these mutant forms of p53 are actually doing is a big question in cancer research.
Prives' team designed experiments to sort this mystery out. By studying cancer cells grown in an artificial system that resembles the three-dimensional structures in the human breast, the researchers learned that cells carrying mutant p53 grow in a disorganized and invasive manner, characteristic of human breast cancers. When the researchers lowered the levels of mutant p53, the 3D cell cultures grew more normally.
Further studies, led by study first author William Freed-Pastor, traced the structural changes to a cholesterol-building pathway (called the mevalonate pathway). This is the same pathway targeted by cholesterol-lowering statins. When the mutant p53 cells were treated with statins, they stopped their disorganized, invasive growth, and in some cases, even died. Importantly, the effects of the drugs were erased when intermediates of the mevalonate pathway were added back in, additional proof that the drug wasn't working in some other, off-target way.
With collaborators in Norway, Prives and Freed-Pastor analyzed breast cancer tissue taken from patients to find that mutations in p53 and elevated activity of mevalonate pathway genes tend to go together in human tumors too. While those findings are encouraging that the cell culture findings may have clinical relevance, Prives emphasizes that it will take considerably more work to confirm that.
"It is what it is," she says. "There are great implications, but nothing clinical yet. Perhaps one could do a clinical trial, and that may support these findings, or it may be more complicated.
Source: Cell Press
Important Gene-Regulation Proteins Pinpointed By New Method

A novel technique has been developed and demonstrated at Penn State University to map the proteins that read and regulate chromosomes -- the string-like structures inside cells that carry genes. The specific order in which these proteins attach DNA-containing nucleosomes along the chromosome determines whether a brain cell, a liver cell, or a cancer cell is formed. Until now, it has been exceedingly difficult to determine exactly where such proteins bind to the chromosome, and therefore how they work. The new technique precisely pinpoints their location, and has the potential to take high-resolution snapshots of proteins as they regulate or miss-regulate an entire genome. The research was published on 18 January 2012 as an Advance Online Publication in the journalNature <www.nature.com>. Related research by the Penn State scientists recently was published in the journal Cell.
The research process, lead by Willaman Professor of Molecular Biology B. Franklin Pugh with Graduate Student Ho Sung Rhee, began by their using a molecular tool called an exonuclease to remove DNA that is not bound by one of the gene-regulating proteins. They then determined the nucleotide sequence for each of the remaining protein-bound DNA bundles -- the sequence of the four major component bases of DNA, labeled A, T, C, and G. "The advantage over other techniques of this technique, called ChIP-exo, is its ability to narrow down any binding location across millions and billions of nucleotide genomes to a certainty of about one nucleotide," Pugh said. "This improvement is roughly analogous to going from a low-resolution 240p television to a high-definition 1080p home-theater system. It provides an unprecedented view into how genes are regulated."
The ChIP-exo technique also removes a substantial amount of noise in the detection system that plagues other methods. The lower-noise technique reveals 2-to-5 times more binding locations, providing a much-more-complete picture of which genes are regulated by a particular protein, as well as a broader understanding of their structural organization across genomes. Having a more-complete picture allows scientists to understand in more detail how gene pathways work in normal human development, or fail to work in disease.
Source: Penn State
Boost for Health? Researchers Isolate Protein Linking Exercise to Health Benefits

A team led by researchers at Dana-Farber Cancer Institute has isolated a natural hormone from muscle cells that triggers some of the key health benefits of exercise. They say the protein, which serves as a chemical messenger, is a highly promising candidate for development as a novel treatment for diabetes, obesity and perhaps other disorders, including cancer.
Bruce Spiegelman, PhD, a cell biologist at Dana-Farber, is senior author of the report, posted as an advanced online publication by the journal Nature. The first author is Pontus Bostroöm, MD, PhD, a postdoctoral fellow in the Spiegelman lab.
"It's exciting to find a natural substance connected to exercise that has such clear therapeutic potential," said Bostroöm.
Spiegelman dubbed the hormone "irisin," after Iris, a Greek messenger goddess. He said the discovery is an important first step in understanding the biological mechanisms that translate physical exercise into beneficial changes throughout the body, both in healthy people and in preventing or treating disease.
"There has been a feeling in the field that exercise 'talks to' various tissues in the body," said Spiegelman, a professor of cell biology at Harvard Medical School. "But the question has been, how?"
According to the report, the irisin hormone has direct and "powerful effects" on adipose, or fatty, tissue -- subcutaneous deposits of white fat that store excess calories and which contribute to obesity.
When irisin levels rise through exercise -- or, in this study, when irisin was injected into mice -- the hormone switches on genes that convert white fat into "good" brown fat. This is beneficial because brown fat burns off more excess calories than does exercise alone.
Only a small amount of brown fat is found in adults, but infants have more -- an evolutionary echo of how mammals keep themselves warm while hibernating. In the wake of findings by Spiegelman and others, there has been a surge of interest in the therapeutic possibilities of increasing brown fat in adults.
Along with stimulating brown fat development, irisin was shown to improve glucose tolerance, a key measure of metabolic health, in mice fed a high-fat diet.
The discovery won't allow people will be able to skip the gym and build muscles by taking irisin supplements, Spiegelman cautioned, because the hormone doesn't appear to make muscles stronger. Experiments showed that irisin levels increase as a result of repeated bouts of prolonged exercise, but not during short-term muscle activity.
The Dana-Farber team identified irisin in a search for genes and proteins regulated by a master metabolic regulator, called PGC1-alpha, that is turned on by exercise. Spiegelman's group had discovered PGC1-alpha in previous research.
Bostroöm said the hunt for molecular targets of increased PGC1-alpha activity ultimately pinpointed irisin, which turned out to be located within the outer membrane of muscle cells. This discovery ran counter to other scientists' contentions that such a protein would reside in the cell's nucleus.
To test whether increasing irisin alone could mimic exercise benefits, the scientists injected modest amounts into sedentary mice that were obese and pre-diabetic.
With 10 days of treatment, the mice had better control of blood sugar and insulin levels -- in effect, preventing the onset of diabetes -- and lost a small amount of weight. Although the weight loss was small, Spiegelman said that the hormone may have a greater effect when given for longer periods.
There were no signs of toxicity or side effects, which was predicted since the researchers limited the increase of irisin to levels typically caused by exercise.
In part because it is a natural substance and because the mouse and human forms of the protein are identical, Spiegelman said it should be possible to move an irisin-based drug rapidly into clinical testing -- perhaps within two years.
The irisin discovery has been licensed by Dana-Farber exclusively to Ember Therapeutics for drug development. Ember is a Boston-based startup co-founded by Spiegelman and scientists at the Joslin Diabetes Center and the Scripps Research Institute in Florida.
The scientists said their findings merely scratch the surface of irisin's multiple effects. They are continuing to explore the hormone's possible benefits in metabolic diseases like diabetes, insulin resistance, and obesity, which constitute a growing epidemic around the world, as well as neurodegenerative illnesses like Parkinson's disease.
Spiegelman added that as growing evidence implicates obesity and physical inactivity in cancer development, it's conceivable irisin-based drugs may have value in prevention and treatment of the disease.
Other authors, in addition to Spiegelman and Boström, are from Dana-Farber; Harvard Medical School; Brigham and Women's Hospital; University of California at San Francisco; Universita Politecnica delle Marche, Ancona, Italy; Odense University Hospital, Denmark; and LakePharma, Belmont, Calif.
The National Institutes of Health funded the research.
Dana-Farber Cancer Institute. "Boost for health? Researchers isolate protein linking exercise to health benefits." ScienceDaily, 11 Jan. 2012. Web. 18 Jan. 2012.
Energy-Saving Chaperon Hsp90

A special group of proteins, the so-called chaperons, helps other proteins to obtain their correct conformation. Until now scientists supposed that hydrolyzing ATP provides the energy for the large conformational changes of chaperon Hsp90. Now a research team from the Nanosystems Initiative Munich could prove that Hsp90 utilizes thermal fluctuations as the driving force for its conformational changes. The renowned journal PNAS reports on their findings. ATP is the major energy source for most organisms and ATPases are the machines, which utilize this fuel, for example to move muscles or cargo in our body. The very abundant chaperone protein Hsp90 has such an ATPase in each of its two monomers. During the last years experiments had suggested that the movement and conformational changes of ATPase proteins are in general strictly linked to ATP binding and hydrolysis (i.e. fuel consumption). To probe this theory Thorsten Hugel, Professor at the Technische Universitaet Muenchen (TUM) and member of the Nanosystems Initiative Munich (NIM), and his team designed a special three color single-molecule FRET (Förster resonance energy transfer) assay with alternating laser excitation (ALEX) for simultaneous observation of ATP binding and conformational changes. Unexpectedly the experiments revealed that binding and hydrolysis of ATP is not correlated with the large conformational changes of Hsp90. Hsp90 is instead a highly flexible machinery driven by thermal fluctuations. "Thermal fluctuations are random changes in the structure of the protein – they can be thought of as collisions with water molecules in the environment, which move rather violently at the temperatures in a living organism," says Thorsten Hugel. "Using these clashes to switch back and forth between different conformations, saves Hsp90 valuable ATP." But then what is the task of ATPase in the Hsp90 chaperone? The scientists suspect that co-chaperones and substrate proteins alter the system so that ATP binding or hydrolysis can take a crucial task. With the newly developed experimental setup, it is now possible to investigate the very complex system in greater detail to resolve this important question. The Munich biophysicists therewith offer a new perspective on the energy conversion in molecular machines.
Source: Technische Universitaet Muenchen
New Insights Into an Ancient Mechanism of Mammalian Evolution

Researchers in the UK have found a simple and widespread way in which DNA is remodeled in six mammalian species, including humans. The study, published in the journal Cell, sheds light on an ancient mechanism of evolution that is still at work in our genome.
A team of geneticists and computational biologists in the UK have reveal how an ancient mechanism is involved in gene control and continues to drive genome evolution. The new study is published in the journal Cell.
To function properly, mammalian tissues require the protein CTCF, which has several key activities including the regulation of genes and interaction with proteins in the cell's nucleus to alter gene activity. CTCF acts by binding to DNA and plays a role in diseases such as HIV infection and cancer. However, very little is known about the origin of the DNA sequences that are bound by CTCF.
In this study, the researchers used samples from six mammals (human, macaque, mouse, rat, dog, and short-tailed opossum) to pinpoint where CTCF binds to each genome. They discovered around 5000 sites that are present in most cell types and tissues, and that have not changed over hundreds of millions of years of mammalian evolution. Because these CTCF binding sites are conserved throughout evolution, the researchers believe that many might play an important role in gene regulation.
The team found an even larger number of locations where CTCF binds DNA in only one lineage or a single species. These additional sites represent a signature of important evolutionary changes since our last common ancestor -- legacies, in some cases, of the evolutionary path to humans. These newer CTCF sites are embedded inside virus-like stretches of DNA called 'retro-transposons'. Retro-transposons use a copy-paste mechanism to spread copies of themselves throughout the genome.
"We developed a new, integrated model of CTCF evolution, which explains the origin of these 5000 highly conserved CTCF binding events in mammals," said Paul Flicek of the European Molecular Biology Laboratory-European Bioinformatics Institute (EMBL-EBI) and the Wellcome Trust Sanger Institute. "Taken together, our findings provide fascinating insight into an ancient mechanism of evolution that is still actively changing our genome."
"CTCF is a key regulator involved in chromatin and gene expression remodelling, both of which are perturbed in the development of cancer. The gene expression and chromatin changes in cancer have also recently been relied on to predict the outcome of specific cancer treatments, which is why it is so important to have a detailed understanding of how particular parts of the genome are resistant or plastic to changes," said Duncan Odom of Cancer Research UK and the Wellcome Trust Sanger Institute.
The retro-transposon's copy-and-paste behaviour has long been considered totally self-serving. However, the study showed that when a retro-transposon containing a CTCF-binding sequence spreads around a mammal's genome, it can deposit functional CTCF binding sites in novel locations, altering the activity of distant genes.
"We looked at six mammalian species representing primates, marsupials, rodents and carnivores, and discovered a simple mechanism that they all use to remodel their DNA," explained Petra Schwalie of EMBL-EBI. "We also found that our distant ancestors also experienced the same complicated relationship between CTCF and retro-transposons."
Using molecular palaeontology techniques, the researchers were able to identify fossil traces of older retro-transposon expansions in the DNA around the shared CTCF binding locations, and showed that this process has been active for hundreds of millions of years.
European Molecular Biology Laboratory. "New insights into an ancient mechanism of mammalian evolution." ScienceDaily, 12 Jan. 2012. Web. 13 Jan. 2012.
Novel Approach to View Inner Workings of Viruses

Since the discovery of the microscope, scientists have tried to visualize smaller and smaller structures to provide insights into the inner workings of human cells, bacteria and viruses. Now, researchers at the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS), part of the National Institutes of Health, have developed a new way to see structures within viruses that were not clearly seen before.
Their findings are reported in the Jan. 13 issue of Science.
Cryo-electron microscopy (cryo-EM) is a technique that allows scientists to image very small particles, like structures on the surface of viruses. This method has been useful in helping researchers understand how vaccines work. But, despite the success of cryo-EM, scientists have been unable to clearly visualize structures inside of viruses, because radiation is used to image them. "With lower doses of radiation, it is not possible to see inside the organism," said lead author Dr. Alasdair Steven of the NIAMS Laboratory of Structural Biology Research. "However, higher doses of radiation damage the virus, destroying the very structures that we would like to view."
Working in collaboration with the group of Dr. Lindsay Black at the University of Maryland Medical School, Baltimore, Steven and his team were able to turn the problem of radiation damage into an asset. Viruses, one of the simplest life forms, are made up of nucleic acids (DNA or RNA) and the proteins encoded by the nucleic acid instruction manual. The researchers realized that proteins inside the virus are more sensitive to damage than DNA.
"We first used low doses of radiation and recorded images in which the inner structure of the virus was invisible," said Steven. "Next, we used high doses of radiation, and found that the inner structure could be seen as a cylinder of bubbles." While the inner structure was damaged, the team was able to superimpose the images, using three-dimensional computer reconstruction. As a result, they were able to clearly visualize the viral structure. The investigators call this technique bubblegram imaging.
Moving forward, the team members anticipate many uses of bubblegram imaging. Ideally, this technique will allow a better understanding of the inner workings of viruses, providing more opportunities for developing novel therapies. Beyond studying viral structure, cryo-EM could be used to visualize interactions of proteins with DNA in human cells. One exciting prospect lies in using this approach to visualize differences in cancer vs. non-cancer cells. "This new cryo-EM procedure renders previously invisible proteins visible and, thus, will provide new understanding of cell biology," said Steven.
NIH/National Institute of Arthritis and Musculoskeletal and Skin Diseases. "Novel approach to view inner workings of viruses."ScienceDaily, 12 Jan. 2012. Web. 13 Jan. 2012.
Receptor for Tasting Fat Identified in Humans

Why do we like fatty foods so much? We can blame our taste buds. Our tongues apparently recognize and have an affinity for fat, according to researchers at Washington University School of Medicine in St. Louis. They have found that variations in a gene can make people more or less sensitive to the taste of fat.
The study is the first to identify a human receptor that can taste fat and suggests that some people may be more sensitive to the presence of fat in foods. The study is available online in the Journal of Lipid Research.
Investigators found that people with a particular variant of the CD36 gene are far more sensitive to the presence of fat than others.
"The ultimate goal is to understand how our perception of fat in food might influence what foods we eat and the quantities of fat that we consume," says senior investigator Nada A. Abumrad, PhD, the Dr. Robert A. Atkins Professor of Medicine and Obesity Research. "In this study, we've found one potential reason for individual variability in how people sense fat. It may be, as was shown recently, that as people consume more fat, they become less sensitive to it, requiring more intake for the same satisfaction. What we will need to determine in the future is whether our ability to detect fat in foods influences our fat intake, which clearly would have an impact on obesity."
People who made more CD36 protein could easily detect the presence of fat. In fact, study subjects who made the most CD36 were eight times more sensitive to the presence of fat than those who made about 50 percent less of the protein.
The researchers studied 21 people with a body mass index (BMI) of 30 or more, which is considered to be obese. Some participants had a genetic variant that led to the production of more CD36. Others made much less. And some were in between.
Participants were asked to taste solutions from three different cups. One contained small amounts of a fatty oil. The other two contained solutions that were similar in texture to the oil but were fat-free. Subjects were asked to choose the cup that was different.
"We did the same three-cup test several times with each subject to learn the thresholds at which individuals could identify fat in the solution," explains first author M. Yanina Pepino, PhD, research assistant professor of medicine. "If we had asked, 'does it taste like fat to you?' that could be very subjective. So we tried to objectively measure the lowest concentration of fat at which someone could detect a difference."
Her team masked input that might help participants identify fat by sight or smell. To eliminate visual cues, they lit the testing area with a red lamp. Study subjects also wore nose clips so that they could not smell the solutions.
Fat is an important component of the diet, and both humans and animals usually prefer high-fat, energy-dense foods. Scientists have believed that people identify those high-fat foods mainly by texture, but this study suggests that the presence of fat can change the way our tongues perceive the food, just as it does for the tastes sweet, sour, bitter, salty and savory (umami).
The CD36 discovery follows research that had identified a role for the gene in rats and mice. Scientists had learned that when animals are genetically engineered without a working CD36gene, they no longer display a preference for fatty foods. In addition, animals that can't make the CD36 protein have difficulty digesting fat.
Up to 20 percent of people are believed to have the variant in the CD36 gene that is associated with making significantly less CD36 protein. That, in turn, could mean they are less sensitive to the presence of fat in food.
Abumrad was the first to identify CD36 as the protein that facilitates the uptake of fatty acids. She says better understanding of how the protein works in people could be important in the fight against obesity.
People with obesity are at an elevated risk for cardiovascular disease, stroke, type 2 diabetes, certain cancers, arthritis and other problems. Obesity rates have risen dramatically over the past 30 years as more people have become sedentary, and diets incorporate more hamburgers, French fries, fried chicken and other high-fat foods.
"Diet can affect sensitivity to fat, and in animals, diet also influences the amount of CD36 that's made," Pepino says. "If we follow the results in animals, a high-fat diet would lead to less production of CD36, and that, in turn, could make a person less sensitive to fat. From our results in this study, we would hypothesize that people with obesity may make less of the CD36 protein. So it would seem logical that the amounts of the protein we make can be modified, both by a person's genetics and by the diet they eat."
Our diet contains fat, mainly in the form of triglycerides, which are made of fatty acids linked to glyerol. In the tast test, the researchers presented subjects with two types of fat. Some cups contained a free fatty acid. Others contained triglyerides.
Pepino and Abumrad knew from animal studies that CD36 is activated by fatty acids but not triglycerides. Human subjects, however, were able to taste both. Pepino believes that's probably due to the activity of an enzyme called lipase in the saliva that breaks the triglycerides, releasing the fatty acids while the fat is still in the mouth.
"Rats, for example, can produce salivary lipase, and the lipase quickly will begin to digest the triglyceride and convert it into a fatty acid," she explains. "In humans, the role of lipase hasn't been as clear. In our experiments, people could detect fat whether it was a triglyceride or a fatty acid."
But when the researchers added the diet drug orlistat, subjects could still taste the fatty acids but were less able to detect the triglycerides. Orlistat inhibits lipase in the mouth, stomach and intestine and is often prescribed to people who are obese to prevent them from absorbing fat in foods.
"Orlistat made it more difficult for people to taste fat," Pepino says. "The solution had to contain higher amounts of triglyceride before they could detect fat. With free fatty acid, however, there was no difference."
Pepino MY, Love-Gregory L, Klein S, Abumrad NA, The fatty acid translocase gene, CD36, and lingual lipase influence oral sensitivity to fat in obese subjects. Journal of Lipid Research, Dec. 31, 2011 [Epub ahead of print].
Washington University School of Medicine. "Blame your taste buds for liking fat: Receptor for tasting fat identified in humans."ScienceDaily, 12 Jan. 2012. Web. 13 Jan. 2012.
Chemotherapy May Influence Leukemia Relapse

The chemotherapy drugs required to push a common form of adult leukemia into remission may contribute to DNA damage that can lead to a relapse of the disease in some patients, findings of a new study suggest.
The research, by a team of physicians and scientists at Washington University School of Medicine in St. Louis, is published Jan. 11 in the advance online edition of Nature.
For patients with acute myeloid leukemia (AML), initial treatment with chemotherapy is essential for putting the cancer into remission. Without it, most patients would die within several months. But even so, about 80 percent of AML patients die within five years when chemotherapy treatment fails to keep the cancer in remission and the disease returns.
Results of the new research provide evidence for a theory that scientists have long held: Chemotherapy contributes to relapse in cancer patients by damaging DNA and generating new mutations that allow tumor cells to evolve and become resistant to treatment.
"The mutations in AML patients who have relapsed are different from those present in the primary tumor, and they are more likely to have a telltale signature of DNA damage," says senior author John F. DiPersio, MD, PhD, the Virginia E. and Sam J. Golman Professor of Medicine and chief of the division of oncology. "This suggests that mutations in the relapse cells are influenced by the chemotherapy drugs the patients receive."
Chemotherapy is known to damage the DNA of both cancer cells and healthy cells. But until now, scientists have had little direct evidence to suggest that chemotherapy itself helps shape the evolution of cancer cells and may contribute to disease recurrence. The researchers suspect this phenomenon is not unique to AML and may occur in other cancers as well.
"Chemotherapy drugs are absolutely necessary to get leukemia patients into remission, but we also pay a price in terms of DNA damage," says co-author Timothy J. Ley, MD, the Lewis T. and Rosalind B. Apple Professor of Oncology. "They may contribute to disease progression and relapse in many different cancers, which is why our long-term goal is to find targeted therapies based on the mutations specific to a patient's cancer, rather than use drugs that further damage DNA."
For the current study, scientists at Washington University's Genome Institute sequenced the genomes – the entire DNA – of cancer cells before and after relapse in eight patients with AML and compared the genetic sequences to healthy cells from the same patients. The data essentially allowed them to map the evolution of cancer cells in each patient.
All the patients received cytarabine and an anthracycline drug to induce remission plus additional chemotherapy in an attempt to keep the cancer from returning. Using technology developed at the Genome Institute, the researchers isolated the DNA segments that contained every mutation in the samples of cancer cells and sequenced those regions nearly 600 times each, far more than the usual 30 times each, which substantially increased the statistical accuracy of the results.
The researchers found that the relapsed cancer cells did not contain a large number of new mutations, as some had predicted. In fact, while the relapsed cells in all the patients had gained some mutations, the percentage was relatively small compared to the number of mutations in the primary tumor.
The scientists also discovered a type of mutation in the relapsed cells that is associated with DNA damage. The frequency of these alterations, known as transversions, was significantly higher for relapse-specific mutations (46 percent) than for primary-tumor mutations (31 percent), suggesting that the chemotherapy may have contributed to some of these mutations, the researchers report. Transversions are also more commonly found in the tumor cells of lung cancer patients who smoke.
Genome sequencing also revealed two major patterns of evolution of cancer cells linked to AML relapse. All patients had a single founding clone: a cluster of cancer cells – all with the same mutations – that define the leukemia. In some patients, the founding clone gains mutations, enabling it to survive chemotherapy and evolve into the relapse clone. In others, a subclone derived from the founding clone survives chemotherapy, gains mutations and evolves to become the dominant clone at relapse.
"It's the same tumor coming back but with a twist," says co-author Richard K. Wilson, PhD, director of the Genome Institute. "It's always the founding clone or a subclone that comes back with new mutations that give the cells new strategies for surviving attack by whatever drugs are thrown at them. This makes a lot of sense but it's been hard to prove without whole-genome sequencing."
In all cases, the chemotherapy failed to kill the founding clone, an indication that eradicating the founding clone and subclones is the key to achieving a cure.
Sequencing the entire genomes of the cancer cells was essential to the researchers' discoveries. Most of the mutations in the relapse samples occurred in the regions of the genome that don't include genes and would have been missed if the researchers had sequenced only a portion of the patients' DNA.
"If we only look at the genes, we typically find a total of 10 to 25 mutations in each patient with AML," says lead author and Genome Institute scientist Li Ding, PhD, research assistant professor of genetics. "That's not enough to see significant changes in the mutational patterns of the primary tumor cells versus those in the relapsed cells. Whole-genome sequencing identifies hundreds of mutations in each patient, which provides the resolution and confidence necessary for us to dig deeper to understand how cancer evolves."
DiPersio, who regularly treats patients with AML, says, "Our preconceived notion of the clonal evolution of AML and other cancers has been altered by our study, which suggests that it is much more complicated and dynamic than we initially suspected and can even be impacted by the therapy that is given to treat the disease."
About 13,000 cases of acute myeloid leukemia are diagnosed each year in the United States. It occurs most often among those age 60 or older and becomes more difficult to treat as patients age. According to the American Cancer Society, the five-year survival rate for AML is 21 percent.
Source: Washington University School of Medicine
UNC Scientists Collaborate To Find First Major Genetic Mutation Associated With Hereditary Prostate Cancer Risk

Chapel Hill - After a 20-year quest to find a genetic driver for prostate cancer that strikes men at younger ages and runs in families, researchers have identified a rare, inherited mutation linked to a significantly higher risk of the disease.
A report on the discovery was published in the January 12, 2012 issue of the New England Journal of Medicine. UNC-Chapel Hill scientist Ethan Lange, PhD, was part of the team of investigators at the Johns Hopkins University School of Medicine, the University of Michigan Health System, Wake Forest University and the Translational Genomics Research Institute.
Lange is associate professor of genetics and biostatistics and a member of UNC Lineberger Comprehensive Cancer Center. The research team found that men who inherit this mutation have a 10 to 20 times higher risk of developing prostate cancer.
Lange explains, "For the first time we have identified an inherited high-risk mutation for prostate cancer. The mutation is significantly more common in men with a family history of prostate cancer that strikes at an earlier age, compared to older patients with no family history. Our findings suggest it could be a valuable early screening tool for men, particularly those with a family history of early-onset disease. The benefit to this population of men could be similar to the benefit of current screening strategies employed for BRCA1 and BRCA2 mutations in women with family history of early-onset breast cancer.
"There is still work to be done regarding understanding the biological function of the mutation and the precise level of absolute risk for carriers of this mutation - a process that took years for the BRCA1 and BRCA2 genes. Still, our results strongly suggest this is the most clinically important mutation identified for prostate cancer to date."
While accounting for only a small fraction of all prostate cancer cases, the discovery may provide important clues about how this common cancer develops and help to identify a subset of men who might benefit from additional or earlier screening. This year, an estimated 240,000 men in the United States will be diagnosed with prostate cancer.
James P. Evans, MD, PhD, Bryson Distinguished Professor of Genetics in the UNC School of Medicine, leads UNC Lineberger's clinical genetics program and is an internationally recognized expert in clinical cancer genetics. He was not involved in the study, but observes, "The genetics of prostate cancer have proven surprisingly difficult to unravel and this work represents significant and welcome progress. While fewer than one percent of Caucasian men carry the described mutation in this particular gene, for those men who do carry it, the increased risk for developing prostate cancer is likely greater than for any previous mutation found to date. Larger follow-up studies will be necessary to understand the importance of this finding for prostate cancer, and it remains to be seen whether this mutation is associated with other cancers."
Lange and Kathleen Cooney, MD, one of two study senior authors, were the first to identify the human chromosome region of interest where the mutation, called HOXB13, was ultimately found. Lange, who has been a collaborator on Dr. Cooney's University of Michigan Prostate Cancer Genetics Project for 17 years, led the statistical analyses and was actively involved in designing the study and interpreting the findings for the current study.
This particular mutation was found in families of European descent, while two different mutations on the HOXB13 gene were identified in families of African descent. Since only seven of the 94 families studied were of African descent, more research will be required before the significance of those mutations is known. African-American men are more likely to be diagnosed with prostate cancer at younger ages and have a more aggressive form of the disease.
Lange concludes, "Over the past couple years, genetic sequence analysis - the ability to evaluate nearly every base of the DNA code over large genomic regions as opposed to a relatively small number of preselected bases - has become possible for large research studies. The National Institutes of Health and private research companies have invested hundreds of millions of dollars into developing this technology, and our results represent one of the first successes demonstrating its use in identifying a strong but uncommon genetic risk factor for a common disease. This new technology should, in the near future, lead to many exciting discoveries of genes important to a wide range of common, complex human diseases."
Source: University of North Carolina School of Medicine
Did An Earlier Genetic Molecule Predate DNA And RNA?

In the chemistry of the living world, a pair of nucleic acids—DNA and RNA—reign supreme. As carrier molecules of the genetic code, they provide all organisms with a mechanism for faithfully reproducing themselves as well as generating the myriad proteins vital to living systems.
Yet according to John Chaput, a researcher at the Center for Evolutionary Medicine and Informatics, at Arizona State University’s Biodesign Institute®, it may not always have been so.
Chaput and other researchers studying the first tentative flickering of life on earth have investigated various alternatives to familiar genetic molecules. These chemical candidates are attractive to those seeking to unlock the still-elusive secret of how the first life began, as primitive molecular forms may have more readily emerged during the planet’s prebiotic era.
One approach to identifying molecules that may have acted as genetic precursors to RNA and DNA is to examine other nucleic acids that differ slightly in their chemical composition, yet still possess critical properties of self-assembly and replication as well as the ability to fold into shapes useful for biological function.
According to Chaput, one interesting contender for the role of early genetic carrier is a molecule known as TNA, whose arrival on the primordial scene may have predated its more familiar kin. A nucleic acid similar in form to both DNA and RNA, TNA differs in the sugar component of its structure, using threose rather than deoxyribose (as in DNA) or ribose (as in RNA) to compose its backbone.
In an article released online today in the journal Nature Chemistry, Chaput and his group describe the Darwinian evolution of functional TNA molecules from a large pool of random sequences. This is the first case where such methods have been applied to molecules other than DNA and RNA, or very close structural analogues thereof. Chaput says “the most important finding to come from this work is that TNA can fold into complex shapes that can bind to a desired target with high affinity and specificity”. This feature suggests that in the future it may be possible to evolve TNA enzymes with functions required to sustain early life forms.
Nearly every organism on earth uses DNA to encode chunks of genetic information in genes, which are then copied into RNA. With the aid of specialized enzymes known as polymerases, RNA assembles amino acids to form essential proteins. Remarkably, the basic functioning of the genetic code remains the same, whether the organism is a snail or a senator, pointing to a common ancestor in the DNA-based microbial life already flourishing some 3.5 billion years ago.
Nevertheless, such ancestors were by this time quite complex, leading some scientists to speculate about still earlier forms of self-replication. Before DNA emerged to play its dominant role as the design blueprint for life, a simpler genetic world dominated by RNA may have prevailed. The RNA world hypothesis as it’s known alleges that ribonucleic acid (RNA) acted to store genetic information and catalyze chemical reactions much like a protein enzyme, in an epoch before DNA, RNA and proteins formed the integrated system prevalent today throughout the living world.
While the iconic double helix of DNA is formed from two complimentary strands of nucleotides, attached to each other by base pairing in a helical staircase, RNA is single-stranded. The two nucleic acids DNA and RNA are named for the type of sugar complex that forms each molecule’s sugar-phosphate backbone—a kind of molecular thread holding the nucleotide beads together.
Could a simpler, self-replicating molecule have existed as a precursor to RNA, perhaps providing genetic material for earth’s earliest organisms? Chaput’s experiments with the nucleic acid TNA provide an attractive case. To begin with, TNA uses tetrose sugars, named for the four-carbon ring portion of their structure. They are simpler than the five-carbon pentose sugars found in both DNA and RNA and could assemble more easily in a prebiotic world, from two identical two-carbon fragments.
This advantage in structural simplicity was originally thought to be an Achilles’ heel for TNA, making its binding behavior incompatible with DNA and RNA. Surprisingly, however, research has now shown that a single strand of TNA can indeed bind with both DNA and RNA by Watson-Crick base pairing—a fact of critical importance if TNA truly existed as a transitional molecule capable of sharing information with more familiar nucleic acids that would eventually come to dominate life.
In the current study, Chaput and his group use an approach known as molecular evolution to explore TNA’s potential as a genetic biomolecule. Such work draws on the startling realization that fundamental Darwinian properties—self-replication, mutation and selection—can operate on non-living chemicals.
Extending this technique to TNA requires polymerase enzymes that are capable of translating a library of random DNA sequences into TNA. Once such a pool of TNA strands has been generated, a process of selection must successfully identify members that can perform a given function, excluding the rest. As a test case, the team hoped to produce through molecular evolution, a TNA strand capable of acting as a high-specificity, high-affinity binding receptor for the human protein thrombin.
They first attempted to demonstrate that TNA nucleotides could attach by complementary base pairing to a random sequence of DNA, forming a hybrid DNA-TNA strand. A DNA polymerase enzyme assisted the process. Many of the random sequences, however, contained repeated sections of the guanine nucleotide, which had the effect of pausing the transcription of DNA into TNA. Once random DNA libraries were built excluding guanine, a high yield of DNA-TNA hybrid strands was produced.
The sequences obtained were 70 nucleotides in length, long enough Chaput says, to permit them to fold into shapes with defined binding sites. The DNA-TNA hybrids were then incubated with the target molecule thrombin. Sequences that bound with the target were recovered and amplified through PCR. The DNA portion was removed and used as a template for further amplification, while the TNA molecules displaying high-affinity, high specificity binding properties were retained.
Additionally, the binding affinity of the evolved and selected TNA molecules was tested against two other common proteins, for which they displayed no affinity, strengthening the case that a highly specific binding molecule had resulted from the group’s directed evolution procedure.
Chaput suggests that issues concerning the prebiotic synthesis of ribose sugars and the non-enzymatic replication of RNA may provide circumstantial evidence of an earlier genetic system more readily produced under primitive earth conditions. Although solid proof that TNA acted as an RNA precursor in the prebiotic world may be tricky to obtain, Chaput points to the allure of this molecule as a strong candidate, capable of storing information, undergoing selection processes and folding into tertiary structures that can perform complex functions. This result provides the motivation to explore TNA as an early genetic system.
Chaput is optimistic that major questions about the prebiotic synthesis of TNA, its role in the origin and early evolution of life on earth, and eventual genetic takeover by RNA will, over time, be answered.
Source: Arizona State University
New Gene, New Mechanism For Neuron Loss In Hereditary Spastic Paraplegias
Hereditary spastic paraplegias (HSPs) are a group of inherited neurodegenerative disorders characterized by progressive weakness and spasticity (stiffness) of the legs. Mutations in more than 30 genes have been linked to HSPs. A team of researchers — led by Stephan Züchner, at the University of Miami Miller School of Medicine, Miami; Evan Reid, at the University of Cambridge, United Kingdom; and Antonio Orlacchio, at the Centro Europeo di Ricerca sul Cervello–Istituto di Ricovero e Cura a Carattere Scientifico Santa Lucia, Italy — has now associated mutations in the gene reticulon 2 with hereditary spastic paraplegia type 12. In addition to identifying a new HSP-associated gene, the team was able to uncover how the mutations in reticulon 2 are likely to cause neurodegeneration, providing new insight into this diverse group of inherited disorders.
Source: Journal of Clinical Investigation
Researchers Map Potential Genetic Origins, Pathways Of Lung Cancer In Never-Smokers
SAN DIEGO -- Researchers have begun to identify which mutations and pathway changes lead to lung cancer in never-smokers — a first step in developing potential therapeutic targets.
Never-smokers (defined as an individual who smoked fewer than 100 cigarettes in his or her lifetime) are estimated to account for 10 percent of lung cancer cases. However, in the past, researchers have not examined this patient population as extensively as they have studied patients with lung cancer who smoked, according to Timothy G. Whitsett, Ph.D., senior postdoctoral fellow in the cancer and cell biology division at the Translational Genomics Research Institute (TGen).
He presented findings on potential gene mutations and pathway alterations that could lead to lung cancer in never-smokers at the AACR-IASLC Joint Conference on Molecular Origins of Lung Cancer: Biology, Therapy and Personalized Medicine, held Jan. 8-11, 2012.
"This is the starting point. We certainly have a lot of pathways and gene expression alterations that we're going to be very interested in confirming and looking at in larger cohorts of patients," Whitsett said. "This is a very important subset of patients with lung cancer, and our research looks to identify pathways and genes that are potentially driving this form of cancer."
Whitsett and his colleagues looked at three female patients with adenocarcinoma: one never-smoker with early-stage disease, one never-smoker with late-stage disease, and, as a comparison, one smoker with early-stage disease. The team performed whole genome sequencing (WGS) and whole transcriptome sequencing (WTS) on each patient to identify gene mutations and pathway alterations that could have led to the development and progression of their specific lung cancers.
"In the never-smoker with early-stage cancer, there were very few mutations in the genome, but when we looked at the whole transcriptome, we saw differences in gene expression," said Whitsett.
In the never-smoker with late-stage disease, the researchers found mutations in what Whitsett called "classic tumor-suppressor genes." He and his colleagues hypothesized that mutations of the tumor-suppressor genes might be a factor in late-stage lung cancer in never-smokers.
Notably, Whitsett and his colleagues reported that these never-smokers' tumors lacked alterations in common genes associated with lung cancer such as EGFR, KRAS and EML/ALK translocations. This finding makes these patients ideal cases for the discovery of new mutations that may drive lung adenocarcinomas in never-smokers, according to the researchers.
Whitsett said that using WGS and WTS to identify cancer origins "has become a way to really dive down into an individual tumor to try to understand the pathways that may be driving that tumor and identify what therapeutic interventions may be possible."
The researchers are now validating these findings in about 30 never-smokers with lung adenocarcinoma and about 60 clinically matched smokers with lung adenocarcinoma.
Source: American Association for Cancer Research
Nanoparticles Hold Promise as Potential Vehicle for Drug Delivery in Brain

In the images of fruit flies, clusters of neurons are all lit up, forming a brightly glowing network of highways within the brain.
It's exactly what University at Buffalo researcher Shermali Gunawardena was hoping to see: It meant that ORMOSIL, a novel class of nanoparticles, had successfully penetrated the insects' brains. And even after long-term exposure, the cells and the flies themselves remained unharmed.
The particles, which are tagged with fluorescent proteins, hold promise as a potential vehicle for drug delivery.
Each particle is a vessel, containing cavities that scientists could potentially fill with helpful chemical compounds or gene therapies to send to different parts of the human body. Gunawardena is particularly interested in using ORMOSIL -- organically modified silica -- to target problems within neurons that may be related to neurodegenerative disorders including Alzheimer's disease.
The recent study on fruit flies is a step toward making this happen, demonstrating that long-term exposure to ORMOSIL, through breathing and feeding, did not injure the animals.
The research appeared in the journal PLoS ONE on Jan. 3.
"We saw that after feeding these nanoparticles in the fruit fly larvae, the ORMOSIL was going mainly into the guts and skin. But over time, in adult flies, you could see it in the brain. These results are really fascinating because these particles do not show any toxic effects on the whole organism or the neuronal cells," said Gunawardena, an assistant professor of biological sciences and a researcher in UB's Institute for Lasers, Photonics and Biophotonics.
The ORMOSIL particles she is investigating are a unique variety crafted by a research group led by Paras N. Prasad, the UB institute's executive director. Each particle contains cavities that can hold drugs, which can be released when the particles are exposed to light.
Besides Gunawardena and Prasad, co-authors on the study include Farda Barandeh, Phuong-Lan Nguyen, Rajiv Kumar, Gary J. Iacobucci, Michelle L. Kuznicki, Andrew Kosterman and Earl J. Bergey, all from UB.
Gunawardena is an expert in axonal transport. This involves the movement of motor proteins along neurons' thread-like axon. These molecular motors, called kinesins and dyneins, carry "cargo" including vital proteins to and from the synapse and cell body of neurons.
In this neuronal highway system, one problem that can occur is an axonal blockage, which resembles a traffic jam in neurons. Proteins aggregate in a clump along the axon.
Researchers don't know whether these obstructions contribute to disorders such as Alzheimer's or Parkinson's diseases, which are characterized by unusual build-ups of proteins called amyloids and Lewy bodies.
But the amyloid precursor protein involved in Alzheimer's disease has been shown to have a role in axonal transport, and if axonal obstructions do turn out to be an early indicator for neurodegeneration seen in Alzheimer's disease, eliminating blockages could help prevent or delay the onset of disease.
That's where ORMOSIL comes in: Gunawardena hopes to use these nanoparticles to target drugs to protein jams along axons, breaking up the accumulations.
Success, if possible, is still a long way off. But the potential benefit is great. Gunawardena calls the research a "high-risk, high-rewards" endeavor.
The next step is for her team to see if they can find a way to force the ORMOSIL to latch onto motor proteins. (The nanoparticles, on their own, do not move along axons.)
University at Buffalo. "Nanoparticles hold promise as potential vehicle for drug delivery in brain." ScienceDaily, 9 Jan. 2012. Web. 10 Jan. 2012.
Tracking Genes' Remote Controls: New Method for Observing Enhancer Activity During Development

As an embryo develops, different genes are turned on in different cells, to form muscles, neurons and other bodily parts. Inside each cell's nucleus, genetic sequences known as enhancers act like remote controls, switching genes on and off. Scientists at the European Molecular Biology Laboratory (EMBL) in Heidelberg, Germany, can now see -- and predict -- exactly when each remote control is itself activated, in a real embryo.
Their work was recently published inNature Genetics.
Stefan Bonn, Robert Zinzen and Charles Girardot, all in Eileen Furlong's lab at EMBL, found that specific combinations of chromatin modifications -- chemical tags that promote or hinder gene expression -- are placed at and removed from enhancers at precise times during development, switching those remote controls on or off.
"Our new method provides cell-type specific information on the activity status of an enhancer and of a gene, within a developing multicellular embryo," says Furlong.
The scientists looked at known enhancers, and compared those that were active to those that were inactive in a type of cells called mesoderm at a particular time in fruit fly development. They noted what chromatin modifications each of those enhancers had, and trained a computer model to accurately predict if an enhancer is active or inactive, based solely on what chromatin marks it bears.
In future, the scientists plan to use this method to study the interplay between the activity status of an enhancer and the presence of key switches, termed transcription factors, at different stages of embryonic development, and in different tissue types, generating an ever more complete picture of how a single cell grows into a complex organism.
European Molecular Biology Laboratory. "Tracking genes' remote controls: New method for observing enhancer activity during development." ScienceDaily, 9 Jan. 2012. Web. 10 Jan. 2012.
World’s First Primate Chimeric Offspring Produced: Research Demonstrates Not All Embryonic Stem Cells Are Equal

Newly published research by scientists at Oregon Health & Science University provides significant new information about how early embryonic stem cells develop and take part in formation of the primate species. The research, which took place at OHSU's Oregon National Primate Research Center, has also resulted in the first successful birth of chimeric monkeys -- monkeys developed from stem cells taken from two separate embryos.
The research is being published this week in the online edition of the journal Cell and will be published in a future printed copy of the journal.
The research was conducted to gain a better understanding of the differences between natural stem cells residing in early embryos and their cultured counterparts called embryonic stem cells. This study also determined that stem cell functions and abilities are different between primates and rodents.
Here's more information about the early primate stem cells that were studied: The first cell type wastotipotent cells -- cells from the early embryo that have the ability to divide and produce all of the differentiated cells in the placenta and the body of organism. These were compared with pluripotent cells -- cells derived from the later stage embryo that have only the ability to become the body but not placenta.
In mice, either totipotent or pluripotent cells from two different animals can be combined to transform into an embryo that later becomes a chimeric animal. However, the current research demonstrated that for reasons yet unknown, chimeric animals can only develop from totipotent cells in a higher animal model: the rhesus macaque. OHSU showed this to be the case by successfully producing the world's first primate chimeric offspring, three baby rhesus macaques named Roku, Hex and Chimero.
"This is an important development -- not because anyone would develop human chimeras -- but because it points out a key distinction between species and between different kind of stem cells that will impact our understanding of stem cells and their future potential in regenerative medicine," explained Shoukhrat Mitalipov, Ph.D., an associate scientist in the Division of Reproductive and Developmental Sciences at ONPRC.
"Stem cell therapies hold great promise for replacing damaged nerve cells in those who have been paralyzed due to a spinal cord injury or for example, in replacing dopamine-producing cells in Parkinson's patients who lose these brain cells resulting in disease. As we move stem cell therapies from the lab to clinics and from the mouse to humans, we need to understand what these cells do and what they can't do and also how cell function can differ in species."
The OHSU Oregon National Primate Research Center and the National Institutes of Health funded the research.
Oregon Health & Science University. "World’s first primate chimeric offspring produced: Research demonstrates not all embryonic stem cells are equal." ScienceDaily, 5 Jan. 2012. Web. 8 Jan. 2012.
Scientists Characterize Protein Essential to Survival of Malaria Parasite

A biology lab at Washington University has just cracked the structure and function of a protein that plays a key role in the life of a parasite that killed 655,000 people in 2010.
The protein is an enzyme thatPlasmodium falciparum, the protozoan that causes the most lethal form of malaria, uses to make cell membrane.
The protozoan cannot survive without this enzyme, but even though the enzyme has many lookalikes in other organisms, people do not make it. Together these characteristics make the enzyme an ideal target for new antimalarial drugs.
The research was published in the January 6 issue of the Journal of Biological Chemistry (JBC) as "Paper of the Week" for that issue.
The work also will be featured inASBMB Today (the newsletter of the American Society for Biological Molecular Biology, which publishes JBC).
Sweating the cold room
The protein's structure might have remained an enigma, had it not been the "unreasonable optimism" of Joseph Jez, PhD, associate professor of biology in Arts & Sciences, which carried his team through a six-year-long obstacle course of failures and setbacks.
"What my lab does is crystallize proteins so that we can see what they look like in three dimensions," Jez says. "The idea is that if we know a protein's structure, it will be easier to design chemicals that would target the protein's active site and shut it down," Jez says.
The lastest discovery is the culmination of a project that began years before when Jez was working at the Danforth Plant Science Center in St. Louis and collaborating with scientists at the local biotech startup Divergence. "At the time, C. eleganshad just been sequenced and the Divergence scientists were looking at using it as an easy model to work out the biochemistry of parasitic nematodes," Jez says.
C. elegans is a free-living nematode, or microscopic roundworm, but many nematodes are parasitic and cause disease in plants, livestock and people.
During this project, Lavanya Palavalli, a summer intern working with Jez, crystallized the C. elegans version of the enzyme. The job of the enzyme, phosphoethanolamine methyltransferase, thankfully abbreviated to PMT, is to add methyl groups to a starting molecule, phosophoethanolamine.
"When Soon Goo Lee later took up the project," says Jez, "the plan was to try to grow better crystals of the C. elegansprotein, ones good enough to get readable X-ray diffraction patterns.
Two years later, the crystals were looking better but still not good enough.
So Jez suggested that Lee go after homologous (look-alike) proteins in other organisms. "Even though the proteins are homologous, each has a different amino acid sequence and so will behave differently in the crystallizations," Jez says. "Lee went from working with two C. elegans proteins to three plant proteins, two other nematode proteins and then thePlasmodium protein," Jez says.
"He took all six of those PMT versions into the crystallization trials to maximize his odds," Jez says.
"To crystallize a protein," Jez says, "we put a solution of a salt or something else that might work as a desiccant in the bottom of a small well. And then we put a drop of our liquid protein on a microscope cover slip and flip it over the top of the well, so the drop of protein is hanging upside down in the well."
"What we're trying to do is to slowly withdraw water from the protein. It's exactly like making rock candy, only in that case, the string hanging into the jar of sugar solution helps to withdraw water," he says.
The difference is that sugar wants to form crystals and proteins are reluctant to do so.
"There are 24 wells to a tray, and we usually screen 500 wells per protein at first," Jez says. "Lee had eight proteins and so his first pass was to screen 4,000 conditions. And then he had to try different combinations of ligands to the proteins and crystallize those. This is why it took a few years to finally get where he needed to go."
Road trip! The scientists need crystals -- preferably nice, big ones -- to stick in the path of an X-ray beam at Argonne National Laboratory in Chicago. (If the crystal is a good one, and all the atoms are lined up in a repeating array, the scattered X-rays will produce a clear pattern of spots.)
Embedded in that pattern is the mathematical information needed to back-calculate to the position of the atoms in the protein, a process a bit like throwing a handful of pebbles in a lake and then calculating where they landed by the pattern of waves arriving at the shoreline.
Lee got the PMT from Haemonchus contortus to crystallize first, but there were technical issues with the diffraction pattern that would have made solving it technically and computationally very demanding.
"When the Plasmodium enzyme finally crystallized, Soon got four crystals kind of stacked on top of each other and each of them was paper thin," Jez says.
"I never thought it would work, but we took them to Argonne anyway and he actually did surgery under the microscope and cracked off a little tiny piece of it."
To everyone's surprise, he got a clean diffraction pattern from the crystal. "Because the Plasmodium enzyme was the smallest one and the easiest to work on, we pushed that one first," Jez says.
The moment of truth "Once we had a Plasmodium crystal that was diffracting really well, we could try back-calculating to see whether we could extract the atom positions from the data," Jez says.
After the computer finished its calculations, Lee clicked a mouse button to see the results, which would reveal whether his years of work finally would pay off.
When Lee clicked the mouse, he got an electron density map in exceptionally sharp focus.
"When you see a map like that, it's like suddenly the wind has kicked up and you're sailing free," Jez says, "because there's this moment, like, before you click that button, no one has ever seen how this protein is put together in three dimensions. You're the first person to ever see it.
"The irony of it is we got such good quality diffraction pattern and electron density maps off such an ugly crystal," he says.
Lock and load "Once you have the electron density map, the task is to build a structure that matches the amino acid sequence of the protein," Jez says.
"The first thing you do is put in the amino acid backbones and connect them together to form a chain. It's like having a long thread, each inch of which is an amino acid, and your job is to take that thread and move it in three dimensions through that electron density map."
The next step is to add the side chains that make one amino acid different from another, Jez says. "The amino acid sequence is known," he says. "Your goal is to match the way you string together the amino acids in the electron density map to that sequence."
"Once you have the overall structure, you can start to figure out how the enzyme works. The PMT enzyme is trying to join two molecules," Jez says. "To do that, it has to lock them in place so that the chemistry can happen, and then it has to let go of them.
"We think the protein has a lid that opens and closes," he says. "The active site stays open until the substrates enter, and then the lid clamps down, and when it clamps down it actually puts the substrates together."
Calling Bill and Melinda Gates Not only do infections byPlasmodium falciparum cause the most severe form of malaria, about 40 percent of the human population lives in areas where the parasite is endemic. Moreover, drugs that used to be effective against malaria are beginning to fail, in part because widespread drug counterfeiting has led to resistance.
New anti-malarial drugs are desperately needed, and the PMT protein is an ideal target. If PMT is disabled, the protozoan can't make cell membranes and it dies. Moreover, a drug that would kill Plasmodium might have minimal side effects on patients.
Although the process of identifying compounds that would target PMT is in the early stages, a handful of anti-parasitical compounds used to treat diseases are known to block PMT as well.
As for Lee, he has had a hard go of it, but now things are breaking his way. Plasmodium PMT is giving up its secrets, and the plant and nematode PMTs are coming along as well.
When he clicked the mouse button and a clean electron density map came up, he says, it was like seeing "the light at the end of a five-year-long tunnel."
Washington University in St. Louis. "Scientists characterize protein essential to survival of malaria parasite." ScienceDaily, 7 Jan. 2012. Web. 8 Jan. 2012.
Scientists Find Structure of Gene-Editing Protein

In the two and a half years since Adam Bogdanove, professor at Iowa State University in the Department of Plant Pathology and Microbiology, along with Matthew Moscou, a former graduate student in that department, discovered how a class of proteins from plant pathogenic bacteria find and bind specific sequences in plant genomes, researchers worldwide have moved fast to use this discovery.
Last year it was first shown that the proteins can be fused to DNA modifying enzymes to manipulate genes and gene functions by Bogdanove and colleagues at the University of Minnesota, led by former ISU professor Dan Voytas, and another group led by Iowa State University faculty member Bing Yang, professor in the Department of Genetics, Development and Cell Biology.
The fused proteins are called TAL effector nucleases, or TALENs, and can be used to better understand gene function in model plant and animal systems, to improve traits in livestock and plants, and even to treat human genetic disorders, according to Bogdanove.
The fact that these proteins can be readily engineered to bind DNA sequences of choice has resulted in a flurry of publications that demonstrate their utility in many different types of cells, including human stem cells.
Largely because of the advent of TALENs, the journal Nature Methods last month named gene editing with engineered nucleases as 2011 Method of the Year.
Now, Bogdanove and researchers from the Fred Hutchinson Cancer Research Center in Seattle have taken the next step by determining the 3-D structure of a TAL effector bound to DNA.
The findings were posted this week on Science Express, a website for early release of papers of exceptional interest that are due to be published in an upcoming issue of the journalScience.
The first author of the study is Amanda Mak, a postdoctoral researcher in the Hutchinson center. Andres Cernadas, a post doctoral researcher in Bogdanove's lab also contributed.
By visualizing the shape of TAL effectors and how they physically interact with the DNA double helix, scientists can now better understand the biochemistry that underlies their ability to recognize and stick to specific DNA sequences.
This will in turn improve scientists' ability to target the proteins to different locations in a genome and to better predict and prevent their binding to unintended, off-target sites, according to Bogdanove.
The structure itself is also interesting from a basic biology standpoint. "It is really quite beautiful," he says, "So far there is nothing else in nature quite like it."
To determine the structure, Bogdanove collaborated with Fred Hutchinson scientists Barry Stoddard, an expert in protein DNA interactions, and Phil Bradley, a computational biologist. Led by Stoddard, the group completed the project in just over a year by using a unique combination of traditional X-ray crystallography and novel computer-based modeling method.
Iowa State University. "Scientists find structure of gene-editing protein." ScienceDaily, 5 Jan. 2012. Web. 8 Jan. 2012.
A Shot of Young Stem Cells Made Rapidly Aging Mice Live Much Longer and Healthier

Mice bred to age too quickly seemed to have sipped from the fountain of youth after scientists at the University of Pittsburgh School of Medicine injected them with stem cell-like progenitor cells derived from the muscle of young, healthy animals. Instead of becoming infirm and dying early as untreated mice did, animals that got the stem/progenitor cells improved their health and lived two to three times longer than expected, according to findings published in the Jan. 3 edition of Nature Communications.
Previous research has revealed stem cell dysfunction, such as poor replication and differentiation, in a variety of tissues in old age, but it's not been clear whether that loss of function contributed to the aging process or was a result of it, explained senior investigators Johnny Huard, Ph.D., and Laura Niedernhofer, M.D., Ph.D. Dr. Huard is professor in the Departments of Orthopaedic Surgery and of Microbiology and Molecular Genetics, Pitt School of Medicine, and director of the Stem Cell Research Center at Pitt and Children's Hospital of PIttsburgh of UPMC. Dr. Niedernhofer is associate professor in Pitt's Department of Microbiology and Molecular Genetics and the University of Pittsburgh Cancer Institute (UPCI).
"Our experiments showed that mice that have progeria, a disorder of premature aging, were healthier and lived longer after an injection of stem cells from young, healthy animals," Dr. Niedernhofer said. "That tells us that stem cell dysfunction is a cause of the changes we see with aging."
Their team examined a stem/progenitor cell population derived from the muscle of progeria mice and found that compared to those from normal rodents, the cells were fewer in number, did not replicate as often, didn't differentiate as readily into specialized cells and were impaired in their ability to regenerate damaged muscle. The same defects were discovered in the stem/progenitor cells isolated from very old mice.
"We wanted to see if we could rescue these rapidly aging animals, so we injected stem/progenitor cells from young, healthy mice into the abdomens of 17-day-old progeria mice," Dr. Huard said. "Typically the progeria mice die at around 21 to 28 days of age, but the treated animals lived far longer -- some even lived beyond 66 days. They also were in better general health."
As the progeria mice age, they lose muscle mass in their hind limbs, hunch over, tremble, and move slowly and awkwardly. Affected mice that got a shot of stem cells just before showing the first signs of aging were more like normal mice, and they grew almost as large. Closer examination showed new blood vessel growth in the brain and muscle, even though the stem/progenitor cells weren't detected in those tissues.
In fact, the cells didn't migrate to any particular tissue after injection into the abdomen.
"This leads us to think that healthy cells secrete factors to create an environment that help correct the dysfunction present in the native stem cell population and aged tissue," Dr. Niedernhofer said. "In a culture dish experiment, we put young stem cells close to, but not touching, progeria stem cells, and the unhealthy cells functionally improved."
Animals that age normally were not treated with stem/progenitor cells, but the provocative findings urge further research, she added. They hint that it might be possible one day to forestall the biological declines associated with aging by delivering a shot of youthful vigor, particularly if specific rejuvenating proteins or molecules produced by the stem cells could be identified and isolated.
Co-authors from the University of Pittsburgh include Mitra Lavasani, Ph.D., Aiping Lu, M.D., and Minjung Song, Ph.D., all of the Stem Cell Research Center and the Department of Orthopaedics; Andria Robinson, of UPCI and Pitt's Graduate School of Public Health; Joseph M. Feduska and Bahar Ahani of the Stem Cell Research Center; Jeremy S. Tilstra, Ph.D., and Chelsea H. Feldman of Pitt's Department of Microbiology and Molecular Genetics; and Paul D. Robbins, Ph.D., of the departments of Orthopaedic Surgery and Microbiology and Molecular Genetics, and UPCI.
The project was funded by grants ES016114, AG033907 and AR051456 from the National Institutes of Health and additional support from The Ellison Medical Foundation, the Henry J. Mankin Endowed Chair at the University of Pittsburgh, and the William F. and Jean W. Donaldson endowed chair at Children's Hospital of Pittsburgh of UPMC.
Source : University of Pittsburgh Schools of the Health Sciences (2012, January 3). A shot of young stem cells made rapidly aging mice live
much
A Firmer Understanding Of Muscle Fibrosis

Researchers describe how increased production of a microRNA promotes progressive muscle deterioration in a mouse model of Duchenne muscular dystrophy (DMD), according to a study published online on January 2 in theJournal of Cell Biology (www.jcb.org).
As DMD patients age, their damaged muscle cells are gradually replaced by collagen-rich, fibrous tissue. This muscle fibrosis is partly induced by the growth factor TGF-beta, which is highly activated in DMD patients, though exactly how this cytokine promotes fibrogenesis is unclear. Pura Muñoz-Cánoves and colleagues examined the role of miR-21, a microRNA whose production is stimulated by TGF-beta signaling.
miR-21 was upregulated in the collagen-producing fibroblasts of both DMD patients and mice that develop disease symptoms similar to human muscular dystrophy (so-called mdx mice). Inhibiting miR-21 reduced collagen levels and prevented, or even reversed, fibrogenesis in diseased animals, whereas mdx mice overexpressing the microRNA produced more collagen and developed fibrotic muscles at earlier ages.
The researchers also discovered that TGF-beta activity and miR-21 production were regulated by the balance of two extracellular factors: uPA—a protease that activates TGF-beta—and its inhibitor PAI-1. mdx mice developed fibrotic muscles more quickly in the absence of PAI-1, but these symptoms could be reversed by inhibiting uPA with a drug or a specific siRNA. In addition to producing more collagen, PAI-1–null fibroblasts also proliferated rapidly because the extra miR-21 induced by active TGF-beta inhibited the tumor-suppressive phosphatase PTEN.
TGF-beta inhibitors prevent muscle fibrosis but have damaging side effects; this study suggests that uPA or miR-21 may make attractive alternative drug targets. Muñoz-Cánoves now wants to investigate the function of miR-21 in other cell types that influence muscle homeostasis, such as the macrophages involved in tissue repair.
Source: Rockefeller University Press
Generex Subsidiary Antigen Express Provides Update On AE37 Cancer Vaccine's Clinical & Regulatory Strategy

WORCESTER, Mass. and TORONTO, Jan. 3, 2012 /PRNewswire/ -- Generex Biotechnology Corporation today provided an update to the clinical development & regulatory strategy for its cancer vaccine, AE37. Positive interim Phase 2b clinical data from the study in HER-2 expressing breast cancer subjects (presented last month at the 34th Annual CTRC-AACR San Antonio Breast Cancer Symposium (SABCS) in San Antonio, Texas) has lead to an earlier-than-expected advancement in the overall development of AE37 for both breast and prostate cancer indications.
AE37 is a novel Ii-Key Hybrid-based HER-2/neu Peptide Vaccine designed to train the immune system, independent of HLA-type, to track down cancer cells throughout the body and destroy them. The positive interim Phase 2b results, building upon completed pre-clinical and Phase I breast cancer studies, enables Antigen Express to move up the timing of key development and regulatory plans. In particular, the company will organize an End-of-Phase 2 meeting with the U.S. Food and Drug Administration (FDA) by the end of Q1 2012. With the FDA's guidance, the Company will move into a pivotal Phase 3 clinical development program in women with loco-regional breast cancer that express low to moderate levels of HER2. Parallel steps will be taken with the European Medicines Agency using the same Phase 2b results. The Phase 2b is currently planned to enroll a total of 300 women and will continue as planned to provide additional valuable information on efficacy and safety of AE37 as the Company pursues its Phase 3 trial plans.
Dr. Eric von Hofe, Ph.D., President of Antigen Express, said: "While the number of patients with recurrent breast cancer is still too low to confirm statistical significance in this ongoing study, the approximate 46% reduction in breast cancer recurrence in low HER2 expressing tumors together with excellent safety enables us to move forward in requesting an End-of-Phase II meeting with the FDA."
Importantly, the majority of women with loco-regional breast cancer have low to moderate expressing HER2 tumors and currently are treated with chemotherapy and radiotherapy only. There is no approved targeted HER2 therapy for these women.
"We are excited and committed to move the AE37 cancer vaccine through the appropriate scientific and regulatory steps," commented Mark Fletcher, President & Chief Executive Officer of Generex. "AE37 not only continues to move closer to market approval but also provides validation for the underlying Ii-Key technology platform upon which it is based."
Phase I studies clearly identified the potential for AE37 to induce the immune system to focus on the HER2 protein through helper (CD4) and cytotoxic (CD8) immune cells in patients. The HER2 protein is expressed on a high percentage of cancers in the breast, prostate, ovaries, gastro-intestinal tract, and lung. In two Phase I trials, AE37 has demonstrated a good safety profile in both breast and prostate cancer patients.
The AE37 cancer vaccine has also completed a Phase 1 trial in prostate cancer demonstrating appropriate dosing and immune activation similar to that seen in the breast cancer trials. The Phase 2b breast trial results add to AE37's database on tolerability and safety and have raised the possibility of using AE37 early in prostate cancer therapy. With the Phase 2b breast cancer trial suggesting immune activation by AE37 could control breast cancer and seeing similar immune activation in the Phase I prostate trial, the Company is encouraged to move AE37 cancer vaccine more rapidly into larger clinical trials in men with newly diagnosed HER2 positive prostate cancer. These Phase 2 trials are being designed with leading oncologists in prostate cancer.
Source: Generex Biotechnology Corporation
Hepatitis C Virus Hijacks Liver MicroRNA

Chapel Hill, NC – Viral diseases are still one of the biggest challenges to medical science. Thanks to thousands of years of co-evolution with humans, their ability to harness the biology of their human hosts to survive and thrive makes them very difficult to target with medical treatment.
Scientists at the University of North Carolina at Chapel Hill, working with colleagues from the University of Colorado, have shown for the first time how a small RNA molecule that regulates gene expression in human liver cells has been hijacked by the hepatitis C virus to ensure its own survival – helping medical scientists understand why a new antiviral drug appears to be effective against the virus.
MicroRNAs are involved in regulating the expression of genes in cells, usually by blocking the production of key proteins or by destabilizing the messenger RNAs that encode the cell's proteins as it grows and divides. Normally they act by downregulating gene expression. The research team found that the binding of a prominent microRNA in liver cells, called miR-122, to the viral RNA results in its stabilization, promoting efficient replication of the virus genome in the liver and supporting the virus' lifecycle.
"The hepatitis C virus has done two very interesting things with miR-122," says Stanley M. Lemon, MD, professor of medicine and microbiology and immunology and a member of UNC Lineberger Comprehensive Cancer Center and the Center for Translational Immunology.
"First, it has evolved a unique relationship with a key regulator, since miR-122 represents about half of all microRNAs present in the liver. Second, the virus has usurped a process that usually downregulates gene expression to upregulate the stability of its RNA and expression of viral proteins needed for its lifecycle. It's a classic example of how viruses subvert normally beneficial functions of the cell to their own nefarious purposes."
Work by Dr. Lemon and his colleagues in 2005 helped to demonstrate that miR-122 was required for hepatitis C to replicate itself, but the mechanism was not understood. Now the UNC research team has shown how it works, which helps to explain how a new experimental antiviral drug target the virus. The drug, called an "antagomer", binds to miR-122 and sequesters it in the liver and thus destabilizes the viral genome, accelerating its degradation in the liver. Results of the most recent study are published online this week in the journal Proceedings of the National Academy of Sciences.
Hepatitis C is a continuing public health problem, which is difficult to measure because symptoms occur months to years after infection. The Centers for Disease Control and Prevention estimates as many as 4 million people in the United States may be persistently infected with hepatitis C virus, and most do not know they are infected. More than a third of those who are long-term carriers may develop chronic liver disease or liver cancer, a deadly form of cancer that is becoming increasingly common due to the spread of this virus.
Source: University of North Carolina School of Medicine
Targeted Therapy Extends Progression-Free Survival Of Patients With Advanced Ovarian Cancer
PHILADELPHIA, PA (December 28, 2011)—Targeted drugs, which block or disrupt particular molecules involved in the growth of tumors, have been shown to be effective treatments against many types of cancer. A new phase 3 clinical trial conducted by the Gynecologic Oncology Group (GOG) showed that a targeted therapy called bevacizumab (Avastin) effectively delayed the progression of advanced ovarian cancer. Patients with newly diagnosed advanced ovarian cancer now typically undergo surgery and chemotherapy, but the new research suggests an additional avenue of treatment. The results of the trial appear in the December 29 issue of the New England Journal of Medicine.
This approach can be looked upon as a third major component of treatment for ovarian cancer and related malignancies," says Robert A. Burger, MD, lead investigator on the GOG study and director of the Women's Cancer Center at Fox Chase Cancer Center. "We've had the combination of surgical management and cytotoxic chemotherapy for many years, but we haven't really seen anything else in terms of a fundamental class of treatment. This represents a new way for us to control the disease."
The placebo-controlled study, which was sponsored by the National Cancer Institute, enrolled 1,873 patients with previously untreated advanced disease from 336 sites, primarily in the United States, but also in Canada, South Korea, and Japan. The patients either had stage III ovarian cancer that could not be entirely removed with surgery, or stage IV disease, and were randomly assigned to one of three groups. For patients who received bevacizumab with chemotherapy followed by bevacizumab for up to an additional 10 months, the median time until their cancer progressed was 14.1 months, compared to 10.3 months for patients in the control group, who received chemotherapy with a placebo and then continued with a placebo. The net effect was a 28% reduction in the risk of disease of ovarian cancer progression over time. Patients who received bevacizumab only with chemotherapy, but not afterward, had a median progression-free survival of 11.2 months.
The National Cancer Institute estimates that nearly 22,000 women were diagnosed with ovarian cancer in 2011, and more than 15,000 died of the disease. For patients diagnosed before the cancer has spread, the five-year relative survival rate is about 93 percent (relative survival measures survival of cancer only, independent of other causes of death). But ovarian cancer is insidious—early symptoms, like bloating, abdominal pain, and trouble eating, are typical of many illnesses and easily dismissed as non-threatening. Women often do not learn they have the disease until it's already spread. In 62 percent of new cases, the patient's cancer has metastasized to distant sites, and the five-year survival rate is just under 27 percent.
Bevacizumab is already FDA-approved for use against some types of colon, lung, kidney and brain cancers; its accelerated approval for metastatic breast cancer was recently revoked by the FDA. The drug acts by binding with vascular endothelial growth factor (VEGF), a protein produced by certain cancers that helps initiate the growth of new blood vessels that feed the tumor. The process of growing new blood vessels is called angiogenesis, and bevacizumab is an angiogenesis inhibitor.
"Bevacizumab blocks the growth factor VEGF, which is important in the process of ovarian cancer progression," says Burger, "and we've seen that this drug is also active in patients with recurrent disease."
Angiogenesis happens at the interface between the host and the disease, which makes it an appealing target for treatment, says Burger, who also led the Phase II GOG study on using bevacizumab in women with recurrent ovarian cancer. He says different ovarian cancers may appear identical under the microscope but differ biologically, which means they'll respond differently to treatment.
In the NEJM paper, Burger and his co-authors point out that another ovarian cancer trial conducted primarily in Europe called ICON7 demonstrated positive results in using becavizumab in combination with chemotherapy and then continued for up to 7 months.
Source: Fox Chase Cancer Center
Gene Identified In Increasing Pancreatic Cancer Risk

PHILADELPHIA — Mutations in the ATM gene may increase the hereditary risk for pancreatic cancer, according to data published in Cancer Discovery, the newest journal of the American Association for Cancer Research.
Pancreatic cancer is one of the most morbid cancers, with less than 5 percent of those diagnosed with the disease surviving to five years. Approximately 10 percent of patients come from families with multiple cases of pancreatic cancer.
"There was significant reason to believe this clustering was due to genetics, but we had not, to this point, been able to find the causative genes that explained the cluster of pancreatic cancer for a majority of these families," said lead author Alison Klein, Ph.D., associate professor of oncology at the Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins and director of the National Familial Pancreas Tumor Registry.
Klein and colleagues used next-generation sequencing, including whole genome and whole exome analyses, and identified ATM gene mutations in two kindreds with familial pancreatic cancer.
When these initial findings were examined in a large series for patients, ATM mutations were present in four of 166 subjects with pancreatic cancer but were absent in 190 spousal control subsets.
Klein said that knowledge of the presence of the ATM gene could lead to better screening for pancreatic cancer, the fourth most common cause of cancer-related death. However, there are currently no recommended screening tests.
Many doctors use endoscopy as a screening tool for pancreatic cancer, but researchers are still evaluating this technique in clinical trials.
Source: American Association for Cancer Research
New Findings About the Prion Protein and Its Interaction With the Immune System

Scrapie is a neurodegenerative disease which can function as a model for other diseases caused by an accumulation of proteins resulting in tissue malformations (proteinpathies), such as Alzheimer's and Parkinson's disease. Many questions regarding these diseases still remain unanswered. A new doctoral study has uncovered a number of factors relating to the uptake of the prion protein (PrPSc) associated with the development of this disease and how this protein interacts with the immune cells in the intestines.
Scrapie in sheep belongs to a group of diseases called "Transmissible spongiform encephalopathies"(TSE) because they are transmitted between individual animals and produce sponge-like, degenerative changes in the brain. These diseases afflict not only sheep but also cattle (BSE), deer (CWD) and humans (CJD). They can to a certain extent also be transmitted between species, as was the case during the 1990s, when over 200 people were infected by food and contracted CJD.
TSE, otherwise known as prion diseases, are thought to be transmitted by means of a diseased variant of a protein, the prion protein, which is a normal component of body cells and is most prolific in the brain. Generally speaking, prion diseases may be infectious, hereditary or occur sporadically/spontaneously. Disease arises when the normal prion protein mutates to the diseased variant, which differs from the healthy prion proteins by its change in structure. The body's cells have difficulty in breaking down this prion protein due to its different structure, and it therefore accumulates.
SInce PrPSc is to be found in the lymphatic tissue of the intestinal system at an early stage of the disease, it is assumed that transmission occurs via the gastrointestinal tract. During her doctoral research, veterinary scientist Caroline Piercey Åkesson studied the uptake of the prion protein in the intestines, thereby throwing new light on processes occurring during the early phase of the disease's development. Contrary to earlier assumptions, she demonstrated by means of immunoelectron microscopy that the prion protein responsible for the disease is not transported directly from the intestines to lymphatic tissue associated with the intestines. On the contrary, she showed that the protein passed freely or in lymphatic cells outside the organised lymphatic tissue in the intestines.
Dendritic cells are presumed to function as "gatekeepers" which determine what the body can tolerate and which immune defence reactions it needs to instigate when confronted with foreign substances. One of the objectives of Åkesson's project was therefore to examine the interaction of dendritic cells with prion protein uptake. Firstly, it was necessary to characterise dendritic cells in healthy sheep intestines and secondly, to investigate which types of cells were associated with the uptake of the prion protein.
Her findings showed that it was not dendritic cells, but macrophages, which were mainly responsible for the uptake of the protein. Åkesson's study revealed that the prion protein makes use of the normal physiological uptake channel for macromolecules in the intestines and that this may have a significant effect on the body's immunological surveillance system. One possible consequence is that immuno-tolerance is stimulated, thus impeding a normal immuno reaction against the prion protein absorbed via the intestines.
Future studies which can reveal how immunological cells are transported and how the prion protein is processed in the body will be of great interest, not only in order to provide more knowledge about scrapie, but also about other neurodegenerative proteinpathies, both in humans and animals.
Cand.med.vet. Caroline Piercey Åkesson defended her doctoral thesis on 20th December 2011 at The Norwegian School of Veterinary Science. The thesis is entitled: Studies on the uptake of prions and their early interaction with immune cells of the sheep gut.
Norwegian School of Veterinary Science (2011, December 29). New findings about the prion protein and its interaction with the immune
Targeted Blocking Of Cell Death Prevents Fatal Condition Septic Shock
Ghent, Belgium 27 December 2011 - Researchers of VIB and UGent have discovered a new approach to preventing septic shock, an often fatal extreme inflammatory reaction of the body. It is the most frequent cause of death at intensive care departments in hospitals. In sepsis, acute inflammation is attended by low blood pressure and blood clots, causing the organs to stop working. Only recently, the Brazilian football legend Socrates, died of the consequences of this condition. In a new study in the top journal Immunity, Peter Vandenabeele and colleagues of VIB-UGent described how blocking a particular form of cell death (necroptosis) fully protects mice against this fatal inflammation.
"This research opens up new perspectives for the treatment of fatal inflammatory diseases such as sepsis," says researcher Peter Vandenabeele of VIB and UGent. "By blocking necroptosis, we have found a possibly new target for a therapy."
Sepsis and SIRS:
The Ghent scientists studied the Systemic Inflammatory Response Syndrome (SIRS). This is a severe inflammatory reaction affecting the entire body. It may be caused by an infection, such as sepsis, or by physical injury such as severe burns or a serious road accident.
Role of TNF in SIRS:
The cytokine tumor necrosis factor (TNF) plays a crucial role in the occurrence of SIRS. The presence of TNF may trigger the cells to cause inflammation and programmed cell death. Inflammation is a necessary response in the body generated, among other things, to prevent or restore damage when injury and infections have been sustained. Programmed cell death can occur in two ways: via apoptosis or via necroptosis. The difference between the two forms of cell death lies among other things in communication with our immune system. Necroptosis usually provokes a strong reaction by the immune system whereas apoptosis proceeds unnoticed.
RIPK: potential therapeutic target for treatment of SIRS and sepsis:
Peter Vandenabeele and his colleagues Linde Duprez, Nozomi Takahashi and Anje Cauwels have discovered that in mice eliminating apoptosis did not have any impact on lethal SIRS whereas eliminating nepcroptosis afforded full protection against the condition. The scientists managed to block nepcroptosis by eliminating RIPK (Receptor-interacting serine/threonine-protein kinase) molecules. The experiments showed that RIPK plays a crucial role in SIRS and sepsis. The molecule appears to constitute a potential therapeutic target for the treatment of SIRS and sepsis. Further research should clarify the potential applications of this discovery.
Source: VIB (the Flanders Institute for Biotechnology)
Mutation In Gene That's Critical For Human Development Linked To Arrhythmia

(SALT LAKE CITY)— Arrhythmia is a potentially life-threatening problem with the rate or rhythm of the heartbeat, causing it to go too fast, too slow or to beat irregularly. Arrhythmia affects millions of people worldwide.
The cardiac conduction system (CCS) regulates the rate and rhythm of the heart. It is a group of specialized cells in the walls of the heart. These cells control the heart rate by sending electrical signals from the sinoatrial node in the heart's right atrium (upper chamber) to the ventricles (lower chambers), causing them to contract and pump blood.
The biologic and genetic mechanisms controlling the formation and function of the CCS are not well understood, but new research with mice shows that altered function of a gene called Tbx3 interferes with the development of the CCS and causes lethal arrhythmias.
In a study published in the Dec. 26, 2011, Proceedings of the National Academy of Sciences early edition, researchers led by the University of Utah showed the CCS is extremely sensitive to levels of Tbx3. Mouse embryos with Tbx3 levels below a critical threshold suffered arrhythmia and couldn't survive. As the levels of Tbx3 were increased, mice survived to birth, but as adults they developed arrhythmias or had sudden death.
Tbx3 dysfunction merits further investigation as a cause of acquired and spontaneous arrhythmias, says Anne M. Moon, M.D., Ph.D., adjunct professor of pediatrics at the U of U School of Medicine and corresponding author on the study. "The cardiac conduction system is very sensitive to Tbx3," Moon says. "Tbx3 is required for the conduction system to develop, mature, and then continue to function properly."
The Tbx3 protein, which is a transcription factor encoded by the TBX3 gene, has been linked to heart development, but its role is not yet clearly defined. Moon and her colleagues, including first author Deborah U. Frank, M.D., Ph.D., U assistant professor of pediatrics, found that slight alterations in the structure of the Tbx3 gene alter the level of the protein in mice. When this happens, it can impair the electrical signal in the sinoatrial node and block the atrioventricular node, which conducts electrical signals from the atria to the ventricles. The result is lethal arrhythmias in embryonic and adult mice.
This discovery has implications for the potential to regenerate functional heart tissue, according to Moon. "There's a big effort to regenerate heart muscle," she says. "But if the muscle can't conduct electrical signals, it's not going to do any good; we also need to be able to regenerate conduction tissues to regulate that muscle."
Arrhythmia is not the first problem related to mutations in the TBX3 gene. In humans, TBX3 mutations have been shown to cause limb malformations in people with ulnar-mammary syndrome, an inherited birth disorder characterized by abnormalities of the bones in the hands and forearms and underdeveloped sweat and mammary glands.
In her future research, Moon wants to discover specifically how Tbx3 regulates the behavior of cells in the cardiac conduction system and whether cells that don't have enough Tbx3 die or turn into some other kind of cells.
"It turns out that Tbx3 is a lot more important in the heart than we realized," Moon says.
Source: University of Utah Health Sciences
Collaborative Effort Uncovers DNA Duplications That May Be Responsible For Genomic-Based Diseases

An important part of saving a species is often understanding its DNA. Through a collaborative effort including 14 scientists representing organizations across Europe and the United States, researchers have been able to analyze the genome of the great ape species of the world.
"A robust appreciation of the means and methods of the evolution of genomes which underlies the diversification of the great apes requires a more detailed knowledge of genome variation that is poorly revealed by current genome sequencing methods. " said Oliver Ryder Ph.D., Director of Genetics for San Diego Zoo Global's Institute of Conservation Research. "This article represents an international collaboration that provides a new level of understanding of the evolutionary dynamics of relatively small DNA duplications that, in humans - and likely great apes as well - may be contributing factors to "genomic" diseases, that in include autism and mental retardation."
The study, published in the August issue of Genome Research, highlights the areas of DNA that appear to be most closely shared by different great ape species. Of particular note is the fact that bonobo and chimpanzee DNA share more copy number variants with gorilla than expected.
Source: Zoological Society of San Diego
Frogs Use Calls To Find Mates With Matching Chromosomes, University Of Missouri Researchers Find

Columbia, MO – When it comes to love songs, female tree frogs are pretty picky. According to a new study from the University of Missouri, certain female tree frogs may be remarkably attuned to the songs of mates who share the same number of chromosomes as they do. The discovery offers insight into how new frog species may have evolved.
Carl Gerhardt, Curators Professor of Biological Sciences in the MU College of Arts and Science and doctoral student Mitch Tucker studied two closely related species of grey tree frogs that live in Missouri, the eastern grey tree frog (Hyla versicolor) and the Cope’s grey tree frog (H. chrysoscelis).
“To the naked eye – human and frog – the two frogs look exactly alike,” Gerhardt said. “The frogs differ only in the number of chromosomes. The eastern grey tree frog has double the number of chromosomes.”
To the ears of potential mates, the two species differ in their vocal performances.
“The males are both singing the same love song – just one frog is singing it slower. It’s kind of like the difference between Eric Clapton’s original and unplugged versions of Layla,” Tucker said.
In previous studies, the scientists found that tree frogs with more sets of chromosome have larger cell sizes, which slows down the trill rate. What was not known was whether the calling preferences of females are similarly linked to chromosome number.
To answer this question, Tucker simulated the chromosome duplication event by replicating spring temperatures early in the frog development. Females were grown to maturity and then exposed to computer-generated, synthetic male calls that differed by trill rate. They found that the females hopped toward the calls with the trill rate of the males with matching chromosome numbers, which indicates female preference.
“This shows that chromosome number alone can control the behavior that keeps the species separate,” Gerhardt said. “In turn, as chromosome number increases, so does the size of cells, which is probably the immediate cause of the changes in calls and preferences.”
In animals, the origin of species is often associated with geographic barriers. A large body of water or range of mountains, for example, splits a large population and prevents mating. The eastern grey tree frog, according to Gerhardt, may represent a rare case of rapid evolution occurring by chromosome duplication, changes in behavior and reproductive isolation.
The report, titled “Parallel changes in mate-attracting calls and female preferences in autotriploid tree frogs,” was published by the journal Proceedings of the Royal Society B-Biological Sciences. The study was supported by funding from the National Science Foundation, National Institutes of Health, and the University of Missouri Research Board.
Source: University of Missouri
Transcriptional Elongation Control Takes On New Dimensions as Researchers Find Gene Class-Specific Elongation Factors

Life is complicated enough, so you can forgive the pioneers of DNA biology for glossing over transcriptional elongation control by RNA polymerase II, the quick and seemingly bulletproof penultimate step in the process that copies the information encoded in our DNA into protein-making instructions carried by messenger RNA.
In a new report appearing in the Dec. 23, 2011, issue of Molecular Cell, researchers at the Stowers Institute for Medical Research add not just a new layer, but a whole new dimension to transcriptional elongation control with evidence that for each class of genes transcribed by RNA polymerase II (Pol II), there exists a specific class of elongation factors.
The Stowers team, led by investigator Ali Shilatifard, Ph.D., discovered that ELL, short for eleven-nineteen lysine-rich leukemia, not only belongs to an assemblage of transcription elongation factors, which Shilatifard's lab had identified as the "Super Elongation Complex" (SEC) a few years ago, but also that ELL is part of a distinct "Little Elongation Complex" (LEC), which acts on a completely different class of genes transcribed by Pol II. Their findings illustrate that the elongation stage of transcription is a much more specific regulator of gene expression than previously believed.
"About fifteen years ago, transcriptional elongation control was not considered all that important for the regulation of gene expression," says Shilatifard of the standard biology textbook descriptions of RNA transcription, which assume that the molecular machinery that supported transcription elongation was one-size-fits-all. "Once RNA polymerase II departed from the promoter regions, it didn't matter all that much what happened next," he says.
Transcriptional elongation is the step following promoter clearance and the step before termination, and was considered to be largely unregulated. The old metaphor was a train running on tracks. "Polymerase is the train. It sat at the promoter -- which would correspond to the station," Shilatifard explains. "The polymerase train would leave the promoter/station and before long would arrive at the end of the gene. The process of the train traveling between the station and the endpoint of the gene -- is considered elongation."
The latest findings derail the train metaphor. "We have shown that there are specific classes of elongation factors for different classes of genes. Therefore, much more is involved than a train simply following a predestined track," he says. "Years ago, B.F. Goodrich (the tire company) advertised that, 'for every road, there is a tire'. What we are learning is that for every class of genes, there seems to be a specific class of elongation factors. The specificity of the complexes seems to control which classes of genes are transcriptionally regulated," says Shilatifard.
Edwin Smith, Ph.D., a research scientist in Shilatifard's lab, identified LEC in Drosophila cells while biochemically dissecting the proteins associated with theDrosophila homolog of the ELL protein. In human cells, where ELL is found within the SEC, it is required to induce the expression of a class of genes specific for the pathogenesis of a subtype of genes involved in acute leukemia.
This type of leukemia results when, through a process known as translocation, the mixed lineage leukemia (MLL) gene becomes fused to any of a number of seemingly unrelated genes. In earlier studies, Shilatifard's group found that many of MLL's fusion partners, including ELL, belong to the SEC. When MLL fuses with any of these unrelated partners, the whole SEC, much like an entourage, now follows MLL to its normal target genes misregulating their elongation and ultimately causing leukemia.
While humans have three ELL genes, fruit flies have only one ELL, but its structural similarity to the human ELLs suggested an evolutionarily conserved and vital function. To find out more about ELL's function in both creatures, Smith searched forDrosophila interaction partners in collaboration with Michael Washburn, Ph.D., and Laurence Florens, Ph.D., who head proteomics at the Stowers. They used their multi-dimensional protein identification technology, or MudPIT, to identify a set of relatively uncharacterized proteins in Drosophila that associate with the C-terminus of ELL in a complex the Shilatifard lab named the "Little Elongation Complex" or LEC.
When Smith knocked down LEC subunits in fruit flies and analyzed the global expression pattern defect with Alexander Garrett, Ph.D., a bioinformatician in the Shilatifard lab, they found that the expression levels of small nuclear RNA (snRNA) genes plummeted. Unlike other RNAs transcribed by RNA pol II, these snRNA molecules are not translated into proteins, instead they team up with proteins to form small nuclear ribonucleoproteins (snRNPs) known by the cheerful name of "snurps." They form the spliceosome, which edits messenger RNA after it is transcribed from DNA. Smith, Garrett, and Chengqi Lin, a graduate student in Shilatifard's laboratory, also demonstrated that this function of LEC is highly conserved fromDrosophila to mammals.
"The specialization of the SEC and LEC complexes for mRNA and snRNA-containing genes, respectively, suggests the presence of specific classes of elongation factors for each class of genes transcribed by RNA polymerase II, which is of fundamental significance," says Smith.
"The next step is to figure out what other classes of genes use other classes of elongation factors. And what are the differential mechanisms of recruitment to RNA Polymerase II on different classes of genes? Once we get a handle on these distinct classes of genes, we hope to be able to modify different classes of genes by modifying these elongation factors." This would be new perspective regarding basic biology and clinical intervention, Shilatifard believes.
Researchers who also contributed to the work include Janet Thornton, Nima Mohaghegh, Deqing Hu, Anita Saraf, Selene K. Swanson, and Christopher Seidel of the Stowers Institute for Medical Research, as well as Jessica Jackson and Joel C. Eissenberg in the Edward A. Doisy Department of Biochemistry and Molecular Biology at the Saint Louis University School of Medicine, Saint Louis, Missouri.
The study was supported in part by the Stowers Institute for Medical Research, and the National Cancer Institute.
Journal Reference:
- Edwin R. Smith, Chengqi Lin, Alexander S. Garrett, Janet Thornton, Nima Mohaghegh, Deqing Hu, Jessica Jackson, Anita Saraf, Selene K. Swanson, Christopher Seidel, Laurence Florens, Michael P. Washburn, Joel C. Eissenberg, Ali Shilatifard. The Little Elongation Complex Regulates Small Nuclear RNA Transcription.Molecular Cell, 2011; 44 (6): 954 DOI:10.1016/j.molcel.2011.12.008
Long Intervening Non-Coding RNAs Play Pivotal Roles in Brain Development

Whitehead Institute scientists have identified conserved, long intervening non-coding RNAs (lincRNAs) that play key roles during embryonic brain development in zebrafish. They also show that the human versions of the lincRNAs can substitute for the zebrafish versions, which implies that the functions of these non-coding RNAs have been retained in humans as well as fish.
Until now, lincRNAs have been studied primarily in cell lines rather than at the organismal level, which has precluded research into how lincRNAs affect growth and development.
"These studies show that zebrafish, an animal that is frequently used to study the genetics of animal development, can also serve as a tool to uncover in systematic fashion the functions of lincRNAs," says Whitehead Member David Bartel, who is also a Howard Hughes Medical Institute investigator and a professor of biology at MIT. "This is another case in which a phenomenon in zebrafish provides insight into what's probably happening in humans, as has been established in many studies of protein-coding genes."
"These studies show that zebrafish, an animal that is frequently used to study the genetics of animal development, can also serve as a tool to uncover in systematic fashion the functions of lincRNAs," says Whitehead Member David Bartel. "This is another case in which a phenomenon in zebrafish provides insight into what's probably happening in humans, as has been established in many studies of protein-coding genes."
Only a minority of RNAs transcribed in a human cell goes on to template protein production, according to a 2007 assessment of the human genome by the Encyclopedia of DNA Elements (ENCODE) Project Consortium, which was funded by the National Human Genome Research Institute. The rest of the RNAs are dubbed non-coding RNAs (ncRNAs), with those located between protein-coding genes and with lengths of 200 base pairs or longer referred to as lincRNAs.
Despite their prevalence in the cell, lincRNAs have been referred to as the "dark matter" of all the transcribed RNAs because little is known of their functions or mechanisms. One limitation to studying this class of RNAs is their low sequence similarity between species. Unlike protein-coding genes, which are frequently well-conserved between species, lincRNA genes typically have a very small bit of conserved DNA between species, if any. This lack of conservation makes identification of related lincRNAs difficult in closely related species and nearly impossible in distantly related species.
For example, Bartel lab scientists Igor Ulitsky and Alena Shkumatava identified more than 500 lincRNAs in zebrafish but found that only 29 of these have homologs in both humans and mice.
Ulitsky and Shkumatava, who report their findings in this week's issue of the journal Cell, tested the function of two of the 29 lincRNAs by knocking them down in zebrafish embryos. Both knockdowns had striking effects on the zebrafish's brain development. Reduction of one of the lincRNAs, which they called cyrano, caused the zebrafish to have enlarged snouts, small heads and eyes, and short, curly tails, while the zebrafish lacking the lincRNA they called megamind had abnormally shaped heads and enlarged brain ventricles.
To test if the human homologs of the cyrano and megamind lincRNAs are functionally equivalent, Shkumatava injected the human versions into the knocked-down zebrafish. Remarkably, the human lincRNAs rescued the zebrafish and restored brain development and head size for both lincRNAs, indicating that the human lincRNAs may have the same role in embryonic development as their zebrafish analogs.
"This work represents a major advance because it provides a framework for studying lincRNAs, a poorly understood, but abundant class of molecules," says Michael Bender, who oversees RNA processing and function grants at the National Institutes of Health's National Institute of General Medical Sciences, which partially funded the work. "The discovery that human lincRNAs appear to function much like their zebrafish counterparts in embryonic development suggests that the framework will prove valuable in bringing new insights on the roles played by lincRNAs in mammalian organisms."
The zebrafish is already a powerful tool for studying genetics. Whitehead Member Hazel Sive, who collaborated with Bartel and his lab members on the Cell paper, uses zebrafish to study brain development and genetic mutations linked to autism.
Says Sive, "The zebrafish is a fantastic, facile system for discovering the mechanisms by which genes work."
"We humans share with zebrafish this subset of ancient, peculiar genes, and the functionality has been retained in them," says Ulitsky. "We can perturb them in zebrafish and then replace them with the human ones and, at least in the lincRNAs we look at, the human ones function to restore proper development."
"Because of this functional conservation of lincRNAs between zebrafish and humans, we're introducing the zebrafish as a new vertebrate tool that could be used basically to uncover the functions of other lincRNAs," says Shkumatava.
This work was supported by the National Institutes of Health's (NIH's) National Institute of General Medical Sciences (NIGMS), European Molecular Biology Organization (EMBO), Human Frontiers Science Program, and the National Science Foundation (NSF).
Journal Reference:
- Igor Ulitsky, Alena Shkumatava, Calvin H. Jan, Hazel Sive, David P. Bartel. Conserved Function of lincRNAs in Vertebrate Embryonic Development despite Rapid Sequence Evolution. Cell, 2011; 147 (7): 1537 DOI:10.1016/j.cell.2011.11.055
Built-In 'Self-Destruct Timer' Causes Ultimate Death of Messenger RNA in Cells

Researchers at Albert Einstein College of Medicine of Yeshiva University have discovered the first known mechanism by which cells control the survival of messenger RNA (mRNA) -- arguably biology's most important molecule. The findings pertain to mRNAs that help regulate cell division and could therefore have implications for reversing cancer's out-of-control cell division. The research was recently described in the journal Cell.
The fate of the mRNA molecules we studied resembles a Greek tragedy," said the study's senior author, Robert Singer, Ph.D., co-director of the Gruss Lipper Biophotonics Center and professor and co-chair of anatomy and structural biology at Einstein. "Their lifespans are determined at the moment of their birth." The study was carried out in yeast cells using advanced microscope technology developed previously by Dr. Singer that has allowed scientists, for the first time, to observe single molecules in single cells in real time.
Directions for making proteins are encoded in the DNA sequences of genes, which reside on chromosomes in the nucleus of each cell. But for proteins to be made, a gene's DNA code must be copied, or transcribed, onto mRNA molecules, which migrate from the nucleus and into the cytoplasm where the cell's protein-making machinery is located. For as long as it exists, an mRNA molecule can act as a template for making copies of a protein. So scientists have long suspected that cells must have ways for degrading mRNAs when, for example, a protein starts accumulating to harmful levels. "The cell somehow decides to destroy its mRNA on cue, but nobody knew how this happens," said Dr. Singer.
In their search for such a mechanism, Dr. Singer and his colleagues focused on two genes, SWI5 and CLB2, which code for proteins that regulate the cell cycle -- the complex series of steps during which a cell divides, first duplicating its genetic material and then distributing it evenly to two daughter cells. To properly choreograph the cell cycle, the levels of the proteins encoded by the SWI5 and CLB2 genes must be exquisitely controlled -- suggesting that the mRNAs made from these genes would be prime candidates for purposeful degradation. Remarkably, the researchers found that these mRNAs are, in effect, born with molecular "self-destruct timers" that ultimately destroy them.
When genes are transcribed, a part of the gene called the promoter region has the job of switching on the gene so that DNA will be copied into mRNA. The Einstein scientists found that the promoter regions of the SWI5 and CLB2 genes do something else as well: they recruit a protein called Dbf2p, which jumps onto mRNA molecules as they're being synthesized.
These mRNAs -- transcribed from the SWI5 and CLB2 genes and bearing the Dbf2p protein -- make their journey from the nucleus into the cytoplasm. Here a protein called Dbf20p joins Dbf2p aboard the mRNA molecules -- and the two proteins together call for the molecules' precipitous decay.
"Our findings indicate that genes making proteins whose levels must be carefully controlled contain promoter regions that sentence their mRNA molecules to death even as the mRNA is being born," said Dr. Singer. "The promoter regions do that by 'marking' the newly made mRNA with the protein Dbf2p -- the common factor between mRNA synthesis and its ultimate decay. Dbf2p stays attached to the mRNA from its birth and then, responding to a signal indicating that no more protein should be made, orders mRNA's destruction."
While these observations pertain to yeast cells, Dr. Singer said he is confident that the process governing mRNA decay in humans "will prove to be very similar" and could be relevant for combating cancer. "Once you gain insight into the mechanisms controlling the cell cycle and cell division," he noted, "you can propose targeted therapies for regulating the uncontrolled cell division that characterizes cancer."
Other scientists involved in the study were lead author Tatjana Trcek (who did this work at Einstein as part of her thesis and is now a postdoctoral student at New York University), Daniel Larson, Ph.D. (now at the National Cancer Institute), postdoctoral fellow Alberto Moldón, Ph.D., and Charles Query, M.D., Ph.D., professor of cell biology at Einstein. This research was supported by the National Institute of General Medical Sciences of the National Institutes of Health.
Journal Reference:
- Tatjana Trcek, Daniel R. Larson, Alberto Moldón, Charles C. Query, Robert H. Singer. Single-Molecule mRNA Decay Measurements Reveal Promoter- Regulated mRNA Stability in Yeast. Cell, 2011; 147 (7): 1484 DOI:10.1016/j.cell.2011.11.051
Possible Cure for Leukemia Found in Fish Oil
A compound produced from fish oil that appears to target leukemia stem cells could lead to a cure for the disease, according to Penn State researchers.
The compound -- delta-12-protaglandin J3, or D12-PGJ3 -- targeted and killed the stem cells of chronic myelogenous leukemia, or CML, in mice, said Sandeep Prabhu, associate professor of immunology and molecular toxicology in the Department of Veterinary and Medical Sciences. The compound is produced from EPA -- Eicosapentaenoic Acid -- an Omega-3 fatty acid found in fish and in fish oil, he said.
"Research in the past on fatty acids has shown the health benefits of fatty acids on cardiovascular system and brain development, particularly in infants, but we have shown that some metabolites of Omega-3 have the ability to selectively kill the leukemia-causing stem cells in mice," said Prabhu. "The important thing is that the mice were completely cured of leukemia with no relapse."
The researchers, who released their findings in the current issue of Blood, said the compound kills cancer-causing stem cells in the mice's spleen and bone marrow. Specifically, it activates a gene -- p53 -- in the leukemia stem cell that programs the cell's own death.
"p53 is a tumor suppressor gene that regulates the response to DNA damage and maintains genomic stability," Prabhu said.
Killing the stem cells in leukemia, a cancer of the white blood cells, is important because stem cells can divide and produce more cancer cells, as well as create more stem cells, Prabhu said.
The current therapy for CML extends the patient's life by keeping the number of leukemia cells low, but the drugs fail to completely cure the disease because they do not target leukemia stem cells, said Robert Paulson, associate professor of veterinary and biomedical sciences, who co-directed this research with Prabhu.
"The patients must take the drugs continuously," said Paulson. "If they stop, the disease relapses because the leukemia stem cells are resistant to the drugs."
Current treatments are unable to kill the leukemia stem cells, Paulson said.
"These stem cells can hide from the treatment, and a small population of stem cells give rise to more leukemia cells," said Paulson. "So, targeting the stem cells is essential if you want to cure leukemia."
During the experiments, the researchers injected each mouse with about 600 nanograms of D12-PGJ3 each day for a week. Tests showed that the mice were completely cured of the disease. The blood count was normal, and the spleen returned to normal size. The disease did not relapse.
In previous experiments, the compound also killed the stem cells of Friend Virus-induced leukemia, an experimental model for human leukemia.
The researchers focused on D12-PGJ3 because it killed the leukemia stem cells, but had the least number of side effects. The researchers currently are working to determine whether the compound can be used to treat the terminal stage of CML, referred to as Blast Crisis. There are currently no drugs available that can treat the disease when it progresses to this stage.
The researchers, who applied for a patent, are also preparing to test the compound in human trials.
Ref:
Penn State (2011, December 22). Possible cure for leukemia found in fish oil. ScienceDaily. Retrieved December 26, 2011, from.
Computer Assisted Design (CAD) for RNA: Researchers Develop CAD-Type Tools for Engineering RNA Control Systems

The computer assisted design (CAD) tools that made it possible to fabricate integrated circuits with millions of transistors may soon be coming to the biological sciences. Researchers at the U.S. Department of Energy (DOE)'s Joint BioEnergy Institute (JBEI) have developed CAD-type models and simulations for RNA molecules that make it possible to engineer biological components or "RNA devices" for controlling genetic expression in microbes. This holds enormous potential for microbial-based sustainable production of advanced biofuels, biodegradable plastics, therapeutic drugs and a host of other goods now derived from petrochemicals.
"Because biological systems exhibit functional complexity at multiple scales, a big question has been whether effective design tools can be created to increase the sizes and complexities of the microbial systems we engineer to meet specific needs," says Jay Keasling, director of JBEI and a world authority on synthetic biology and metabolic engineering. "Our work establishes a foundation for developing CAD platforms to engineer complex RNA-based control systems that can process cellular information and program the expression of very large numbers of genes. Perhaps even more importantly, we have provided a framework for studying RNA functions and demonstrated the potential of using biochemical and biophysical modeling to develop rigorous design-driven engineering strategies for biology."
Keasling, who also holds appointments with the Lawrence Berkeley National Laboratory (Berkeley Lab) and the University of California (UC) Berkley, is the corresponding author of a paper in the journal Science that describes this work. The paper is titled "Model-driven engineering of RNA devices to quantitatively-program gene expression." Other co-authors are James Carothers, Jonathan Goler and Darmawi Juminaga.
Synthetic biology is an emerging scientific field in which novel biological devices, such as molecules, genetic circuits or cells, are designed and constructed, or existing biological systems, such as microbes, are re-designed and engineered. A major goal is to produce valuable chemical products from simple, inexpensive and renewable starting materials in a sustainable manner. As with other engineering disciplines, CAD tools for simulating and designing global functions based upon local component behaviors are essential for constructing complex biological devices and systems. However, until this work, CAD-type models and simulation tools for biology have been very limited.
Identifying the relevant design parameters and defining the domains over which expected component behaviors are exerted have been key steps in the development of CAD tools for other engineering disciplines," says Carothers, a bioengineer and lead author of the Science paper who is a member of Keasling's research groups with both JBEI and the California Institute for Quantitative Biosciences. "We've applied generalizable engineering strategies for managing functional complexity to develop CAD-type simulation and modeling tools for designing RNA-based genetic control systems. Ultimately we'd like to develop CAD platforms for synthetic biology that rival the tools found in more established engineering disciplines, and we see this work as an important technical and conceptual step in that direction."
Keasling, Carothers and their co-authors focused their design-driven approach on RNA sequences that can fold into complicated three dimensional shapes, called ribozymes and aptazymes. Like proteins, ribozymes and aptazymes can bind metabolites, catalyze reactions and act to control gene expression in bacteria, yeast and mammalian cells. Using mechanistic models of biochemical function and kinetic biophysical simulations of RNA folding, ribozyme and aptazyme devices with quantitatively predictable functions were assembled from components that were characterized in vitro,in vivo and in silico. The models and design strategy were then verified by constructing 28 genetic expression devices for theEscherichia coli bacterium. When tested, these devices showed excellent agreement -- 94-percent correlation -- between predicted and measured gene expression levels.
"We needed to formulate models that would be sophisticated enough to capture the details required for simulating system functions, but simple enough to be framed in terms of measurable and tunable component characteristics or design variables," Carothers says. "We think of design variables as the parts of the system that can be predictably modified, in the same way that a chemical engineer might tune the operation of a chemical plant by turning knobs that control fluid flow through valves. In our case, knob-turns are represented by specific kinetic terms for RNA folding and ribozyme catalysis, and our models are needed to tell us how a combination of these knob-turns will affect overall system function."
JBEI researchers are now using their RNA CAD-type models and simulations as well as the ribozyme and aptazyme devices they constructed to help them engineer metabolic pathways that will increase microbial fuel production. JBEI is one of three DOE Bioenergy Research Centers established by DOE's Office of Science to advance the technology for the commercial production of clean, green and renewable biofuels. A key to JBEI's success will be the engineering of microbes that can digest lignocellulosic biomass and synthesize from the sugars transportation fuels that can replace gasoline, diesel and jet fuels in today's engines.
"In addition to advanced biofuels, we're also looking into engineering microbes to produce chemicals from renewable feedstocks that are difficult to produce cheaply and in high yield using traditional organic chemistry technology," Carothers says.
While the RNA models and simulations developed at JBEI to date fall short of being a full-fledged RNA CAD platform, Keasling, Carothers and their coauthors are moving towards that goal.
"We are also actively trying to make our models and simulations more accessible to researchers who may not want to become RNA control system experts but would nonetheless like to use our approach and RNA devices in their own work," Carothers says.
While the work at JBEI focused on E. coli and the microbial production of advanced biofuels, the authors of the Sciencepaper believe that their concepts could also be used for programming function into mammalian systems and cells.
"We recently initiated a research project to investigate how we can use our approach to engineer RNA-based genetic control systems that will increase the safety and efficacy of regenerative medicine therapies that use cultured stem cells to treat diseases such as diabetes and Parkinson's," Carothers says.
This research was supported in part by grants from the DOE Office of Science through JBEI, and the National Science Foundation through the Synthetic Biology Engineering Research Center (SynBERC).
Journal Reference:
- James M. Carothers, Jonathan A. Goler, Darmawi Juminaga, Jay D. Keasling. Model-Driven Engineering of RNA Devices to Quantitatively Program Gene Expression. Science, December 2011: Vol. 334 no. 6063 pp. 1716-1719 DOI: 10.1126/science.1212209
mirEX: a platform for comparative exploration of plant pri-miRNA expression data

Abstract
mirEX is a comprehensive platform for comparative analysis of primary microRNA expression data. RT–qPCR-based gene expression profiles are stored in a universal and expandable database scheme and wrapped by an intuitive user-friendly interface. A new way of accessing gene expression data in mirEX includes a simple mouse operated querying system and dynamic graphs for data mining analyses. In contrast to other publicly available databases, the mirEX interface allows a simultaneous comparison of expression levels between various microRNA genes in diverse organs and developmental stages. Currently, mirEX integrates information about the expression profile of 190 Arabidopsis thaliana pri-miRNAs in seven different developmental stages: seeds, seedlings and various organs of mature plants. Additionally, by providing RNA structural models, publicly available deep sequencing results, experimental procedure details and careful selection of auxiliary data in the form of web links, mirEX can function as a one-stop solution for Arabidopsis microRNA information. A web-based mirEX interface can be accessed at http://bioinfo.amu.edu.pl/mirex.
Schizophrenia: Small genetic changes pose risk for disease

(Medical Xpress) -- Carrying single DNA letter changes from two different genes together may increase the risk of developing schizophrenia, Johns Hopkins researchers reported in the November 16 issue of Neuron.
Causes for psychiatric diseases like schizophrenia and autism have been difficult to pinpoint, since they may be triggered by many small genetic changes that alone may be insufficient, but in the right combination may cause disease.
Drastic DNA rearrangements in the genetic letters of the DISC1 gene are known to cause schizophrenia and other major mental disorders, however, these large changes are rare and do not apply to the majority of people with schizophrenia. Nevertheless, DISC1 is thought to be an entry point for study into the cause of the disease, and defects in DISC1 combined with defects in other genes may contribute to disease.
“We studied the function of two proteins known to interact, FEZ1 and DISC1, in cells and animal models, which suggested that these proteins work together in adult brain development” says Guo-li Ming, M.D., Ph.D., professor of neurology and neuroscience and member of the Johns Hopkins Institute for Cell Engineering. “When we looked at the human genetic sequences of DISC1 and FEZ1, we found that a combination of small DNA changes raises risk for schizophrenia.”
To determine if FEZ1 and DISC1 work together in adult brain development, the researchers used molecular biology techniques to reduce the amount of FEZ1 in the newborn neurons in mouse adult hippocampus, then examined the cells under a microscope. The neurons with less FEZ1 looked similar to cells with less DISC1; they were larger and had longer feelers that are used to reach out and communicate with other neurons nearby. The researchers proposed that these proteins may be working together in neurons to control cell size and feeler length, and when this process is disrupted, it may lead to psychiatric diseases.
Then, the researchers checked existing cases of schizophrenia to see if combinations of single-letter DNA changes in DISC1 and FEZ1 made people more susceptible to the disease. The researchers examined a large patient database, the Genetic Association Information Network, created by the National Institutes of Health to identify genome associated diseases. Using statistical approaches, the researchers examined four different single-letter DNA changes in the FEZ1 sequence from 1,351 schizophrenia cases and 1,378 healthy people. Single-letter DNA changes in FEZ1 alone did not contribute to schizophrenia risk. However, when the researchers looked at these four different FEZ1 DNA letter changes in combination with a DISC1 single DNA letter change already known to slightly increase schizophrenia risk, they found that one particular FEZ1 DNA change along with the DISC1 change significantly increased the risk of schizophrenia by two and a half times.
“By continuing to examine interactions of key genes involved with disease in cells and correlating the results with patient databases, we can begin to unravel the genetic contributions of psychiatric disorders that previously were a mystery to us,” says Hongjun Song, Ph.D., professor of neurology and director of the Stem Cell Program at the Institute for Cell Engineering. “Finding sets of proteins, like FEZ1 and DISC1, that synergistically work together to cause disease will also give us new drug targets to develop new therapies.”
Provided by Johns Hopkins University (news : web)